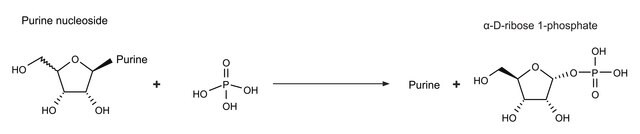
N9914
Polynucleotide phosphorylase from Synechocystis sp.
recombinant, expressed in E. coli
Synonyme(s) :
PNPase, Polyribonucleotide Nucleotidyltransferase
About This Item
Produits recommandés
Source biologique
bacterial (Synechocystis sp.)
Niveau de qualité
Produit recombinant
expressed in E. coli
Description
Histidine tagged
Pureté
90% (SDS-PAGE)
Forme
solution
Activité spécifique
≥500 units/mg protein
Poids mol.
85 kDa
Technique(s)
cell based assay: suitable
Adéquation
suitable for molecular biology
Application(s)
cell analysis
Conditions d'expédition
dry ice
Température de stockage
−70°C
Vous recherchez des produits similaires ? Visite Guide de comparaison des produits
Description générale
Application
Actions biochimiques/physiologiques
Définition de l'unité
Code de la classe de stockage
12 - Non Combustible Liquids
Classe de danger pour l'eau (WGK)
WGK 1
Point d'éclair (°F)
Not applicable
Point d'éclair (°C)
Not applicable
Certificats d'analyse (COA)
Recherchez un Certificats d'analyse (COA) en saisissant le numéro de lot du produit. Les numéros de lot figurent sur l'étiquette du produit après les mots "Lot" ou "Batch".
Déjà en possession de ce produit ?
Retrouvez la documentation relative aux produits que vous avez récemment achetés dans la Bibliothèque de documents.
Les clients ont également consulté
Notre équipe de scientifiques dispose d'une expérience dans tous les secteurs de la recherche, notamment en sciences de la vie, science des matériaux, synthèse chimique, chromatographie, analyse et dans de nombreux autres domaines..
Contacter notre Service technique