MBD0013
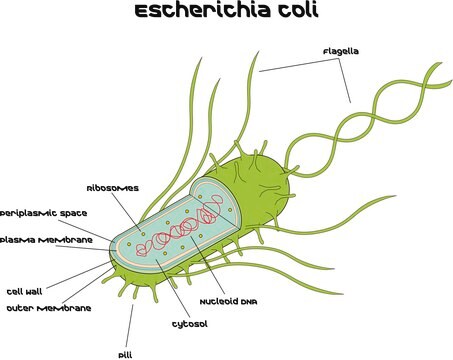
Microbial DNA standard from Escherichia coli
Suitable for PCR, sequencing and NGS, 10 ng/μL
Sinônimo(s):
E.Coli
About This Item
Produtos recomendados
Nível de qualidade
forma
liquid
concentração
10 ng/μL
técnica(s)
DNA extraction: suitable
DNA sequencing: suitable
PCR: suitable
adequação
suitable for restriction endonuclease digests, PCR amplification, Southern blots, and sequencing reactions
Condições de expedição
ambient
temperatura de armazenamento
−20°C
Descrição geral
Read here how to use our standards to ensure data integrity for your microbiome research.
Aplicação
Suitable for Quantitative standard for PCR, Sequencing and NGS
Características e benefícios
- Individual microbial standard for microbiomics and meta-genomics workflow
- Suitable standard for PCR, sequencing and NGS
- Improve Bioinformatics analyses
- Increases reproducibility
- Compare results lab to lab
forma física
Outras notas
Código de classe de armazenamento
12 - Non Combustible Liquids
Classe de risco de água (WGK)
WGK 1
Ponto de fulgor (°F)
Not applicable
Ponto de fulgor (°C)
Not applicable
Certificados de análise (COA)
Busque Certificados de análise (COA) digitando o Número do Lote do produto. Os números de lote e remessa podem ser encontrados no rótulo de um produto após a palavra “Lot” ou “Batch”.
Já possui este produto?
Encontre a documentação dos produtos que você adquiriu recentemente na biblioteca de documentos.
Os clientes também visualizaram
Artigos
An overview of human microbiome research, workflow challenges, sequencing, library production, data analysis, and available microbiome reagents to support your research.
Nossa equipe de cientistas tem experiência em todas as áreas de pesquisa, incluindo Life Sciences, ciência de materiais, síntese química, cromatografia, química analítica e muitas outras.
Entre em contato com a assistência técnica