Detection and Molecular Identification of Mycobacteria Species
Anandi Martin, PhD.
Institute of Tropical Medicine, Mycobacteriology Unit, Belgium
Microbiology Focus Edition 2.2
The diseases produced by the genus Mycobacterium are important causes of morbidity and mortality in the world. The identification of mycobacteria at the species level is important because of the clinical significance as some species are pathogenic while others are not.

Figure 1.A typical colony of Mycobacterium tuberculosis seen under a microscope
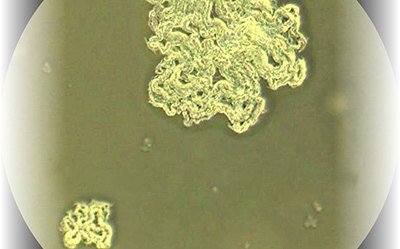
Figure 2.Mycobacterium tuberculosis with 10x magnification
Microbiology and Characteristics of the Genus Mycobacteria
Mycobacteria belong to the order Actinomycetales and are the only genus in the family Mycobacteriaceae. Currently, the genus Mycobacterium has more than 100 recognized or proposed species, including numerous pathogens and saprophytic organisms of warm-blooded animals. Mycobacteria are slender, non-spore-forming, rod-shaped, aerobic, slow-growing, and free-living in soil and water. These bacteria have a generation time of about 20 h, thus isolation and identification may take up to 6 weeks.
Traditionally, mycobacteria are identified by phenotypic methods, based on culture, such as morphological characteristics, growth rates, preferred growth temperature, pigmentation, and a series of biochemical tests. Testing is laborious, difficult, and time-consuming – takes up to several weeks for adequate growth, and sometimes misidentification may occur because different species may have indistinguishable morphological and biochemical profiles. Different culture media are used for the isolation of mycobacteria. The most common are egg-based media known as “Löwenstein-Jensen medium” and contain high concentrations of malachite green to overcome contamination with other bacteria.
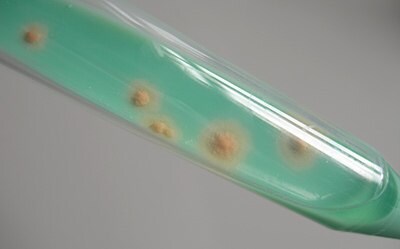
Figure 3.Mycobacteria colonies on TB-Medium

Figure 4.Different mycobacteria species grown on TB-Medium
Culture media for Isolation of Mycobacteria
In the last decade, several commercial systems for mycobacterial culture based on liquid media have been introduced. Liquid culture media is proven to be significantly more sensitive than egg-based solid media for the isolation of mycobacteria from clinical specimens. M. tuberculosis bacilli are slow-growing mycobacteria which means that in primary isolation they hardly show any visible growth during the first week of culture. On egg-based media, they produce characteristic non-pigmented colonies, with a generally rough and dry appearance simulating breadcrumbs. On agar-based media, the colonies appear flat, dry, and rough with irregular edges. M. tuberculosis is niacin positive, inhibited by p-nitrobenzoic acid, and displays nitratase activity. Additional tests that confirm an isolate as M. tuberculosis are susceptible to pyrazinamide when grown on thiophene carboxylic acid hydrazide (TCH), without iron uptake and catalase production at 68 °C.
Detection of mycobacteria using a staining technique
The Ziehl-Neelsen staining for the direct detection of mycobacteria by microscopy is used to identify acid-fast bacilli. The lipid-rich cell wall of mycobacteria makes it resistant to Gram stain. It can also be used to stain a few other bacteria like Nocardia. The reagents used for the staining are carbolfuchsin, acid-alcohol, and methylene blue. Acid-fast bacilli appear bright red after staining.
Non-tuberculous mycobacteria
Non-tuberculous mycobacteria (NTM) are ubiquitous organisms that are frequently isolated from environmental sources, including surface water, tap water, and soil. The NTM species most frequently associated with pulmonary disease are M. avium, M. kansasii and M. abscessus. Injury cutaneous/ subcutaneous infections have been attributed to rapidly growing mycobacteria. M. fortuitum, M. abscessus, and M. chelonae are thought to be caused by local environmental strains or contaminated commercial surgical materials, devices, or solutions for injection. Rapidly growing mycobacteria often grow on classical bacterial culture media, especially on blood agar plates, however, due to their delay in forming visible colonies (up to 10 days), they are usually not detected in the routine bacteriology laboratory. NTM can also be isolated on most media suitable for the isolation of mycobacteria. Although the optimum temperature for most species is 30-32 °C, NTM also grows at 36-37 °C, the standard temperature for isolation of M. tuberculosis.
Identification of mycobacteria species by molecular methods
In the last decade, advances in molecular methods have facilitated the rapid and reliable identification of many mycobacterial species. Nucleic acid probes, species-specific PCR, reverse hybridization, and 16S rRNA sequencing have been evaluated for application in clinical laboratories. The first commercially available method was the AccuProbe (Gen- Probe Inc.), based on species-specific DNA probes that hybridize to rRNA for the identification of several important mycobacteria, including the M. tuberculosis complex, M. avium, M. intracellulare, M. avium complex, M. kansasii, and M. gordonae.
More recently, other molecular commercial systems have also been introduced for the rapid identification of M. tuberculosis complex such as the INNO- LiPA MYCOBACTERIA v2 (Innogenetics NV, Ghent, Belgium), and the GenoType MTBC and GenoType Mycobacterium (Hain Lifesciences, Nehren, Germany).
- INNO-LiPA MYCOBACTERIA v2 is a line probe assay that simultaneously detects and identifies the genus Mycobacterium and 16 different mycobacterial species. It is based on nucleotide differences in the 16S-23S rRNA gene spacers.
- The GenoType MTBC and GenoType Mycobacterium are also based on the reverse line probe hybridization assay and are intended for the differentiation of M. tuberculosis complex members and the identification of 35 species of mycobacteria including M. tuberculosis. The GenoType MTBC is based on a 23S rRNA gene fragment specific for the M. tuberculosis complex, along with gyrB sequence polymorphisms, and the RD1 deletion sequence for identification of M. bovis BCG.
- Several ‘in-house’ techniques are also available with sequencing of the 16S rRNA gene as the reference standard to which all other new techniques are generally compared.
References
To continue reading please sign in or create an account.
Don't Have An Account?