03353575910
Roche
DIG Oligonucleotide 3′-End Labeling Kit, 2nd generation
sufficient for 25 labeling reactions (100 pmol of oligonucleotides per assay; 1 ug of a 30-mer oligonucleotide), storage condition avoid repeated freeze/thaw cycles
About This Item
Recommended Products
usage
sufficient for 25 labeling reactions (100 pmol of oligonucleotides per assay; 1 ug of a 30-mer oligonucleotide)
Quality Level
manufacturer/tradename
Roche
greener alternative product characteristics
Designing Safer Chemicals
Learn more about the Principles of Green Chemistry.
sustainability
Greener Alternative Product
greener alternative category
, Aligned
shipped in
dry ice
General description
Application
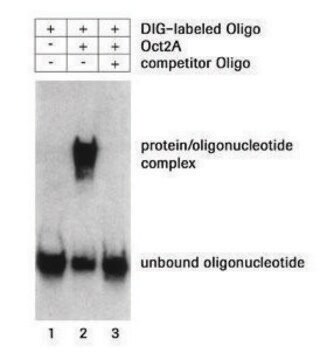
DIG-labeled oligonucleotides has been used in a variety of hybridization techniques:
- dot/slot blots
- colony/ plaque hybridizations
- Southern blots/ northern blots
- in situ hybridizations
Features and Benefits
- Fast hybridization kinetics, due to the small size of oligonucleotides
- Single-stranded probes, no renaturation during hybridization
- Sequence can be designed according to the experiment
- Specially suited for in situ hybridization; due to their small size, the oligonucleotides readily diffuse into fixed tissues and cells
Packaging
Quality
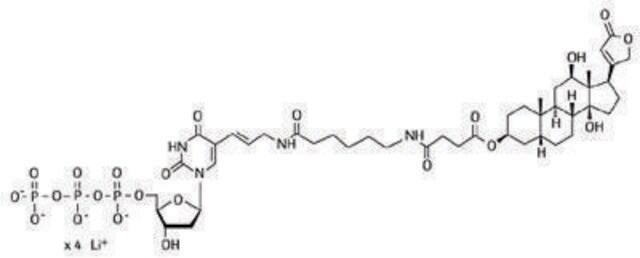
Principle
Preparation Note
Working concentration: Oligonucleotides: 100 pmol
Up to 100 pmol (1 μg of a 30-mer) oligonucleotide can be labeled in a single standard labeling reaction.
Sample Materials
Oligonucleotides of a length from 14 to 100 nucleotides, purified by HPLC or gel electrophoresis
Storage and Stability
Other Notes
Kit Components Only
- Reaction Buffer 5x concentrated
- CoCl<sub>2</sub> Solution 25 mM
- DIG-ddUTP Solution 1 mM
- Recombinant Terminal Transferase 400 U/μl
- Control Oligonucleotide, unlabeled 20 pmol/μl
- Oligonucleotide, DIG-ddUTP labeled 2.5 pmol/μl
- Control DNA, 2.5 pmol/μl pUC 18 DNA, supercoiled
- Glycogen Solution 20 mg/ml
- DNA Dilution Buffer, 50 μg/ml fish sperm DNA
Signal Word
Danger
Hazard Statements
Precautionary Statements
Hazard Classifications
Acute Tox. 4 Inhalation - Acute Tox. 4 Oral - Aquatic Chronic 2 - Carc. 1B Inhalation - Repr. 1B
Storage Class Code
6.1D - Non-combustible, acute toxic Cat.3 / toxic hazardous materials or hazardous materials causing chronic effects
WGK
WGK 3
Flash Point(F)
does not flash
Flash Point(C)
does not flash
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Articles
Digoxigenin (DIG) labeling methods and kits for DNA and RNA DIG probes, random primed DNA labeling, nick translation labeling, 5’ and 3’ oligonucleotide end-labeling.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service