41719
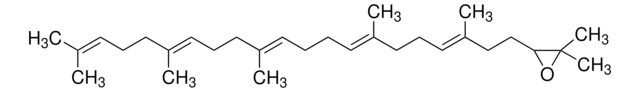
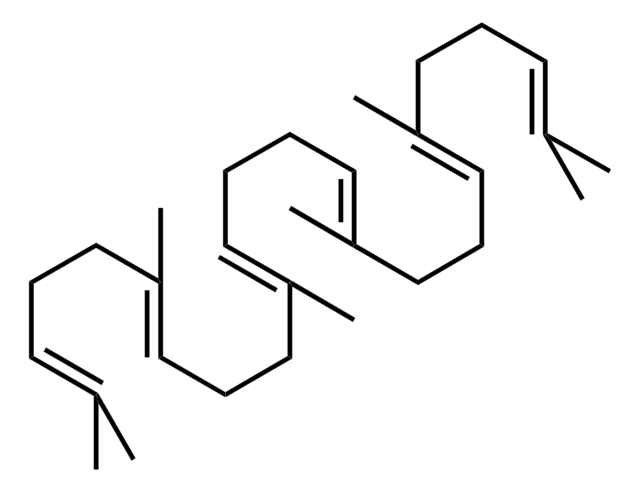
(3S)-2,3-Oxidosqualene
≥97.5% (HPLC)
Sinónimos:
(S)-2,3-Epoxy-2,3-dihydrosqualene, (S)-22,23-Epoxy-2,6,10,15,19,23-hexamethyl-2,6,10,14,18-tetracosapentaene, (S)-Squalene 2,3-epoxide, (S)-Squalene 2,3-oxide
About This Item
Productos recomendados
assay
≥97.5% (HPLC)
form
liquid
optical purity
enantiomeric excess: ≥90.0%
suitability
conforms to structure for Proton NMR spectrum
storage temp.
−20°C
SMILES string
CC1(C)O[C@H]1CC/C(C)=C/CC/C(C)=C/CC/C=C(C)/CC/C=C(C)/CCC=C(C)C
InChI
1S/C30H50O/c1-24(2)14-11-17-27(5)20-12-18-25(3)15-9-10-16-26(4)19-13-21-28(6)22-23-29-30(7,8)31-29/h14-16,20-21,29H,9-13,17-19,22-23H2,1-8H3/b25-15+,26-16+,27-20+,28-21+/t29-/m0/s1
InChI key
QYIMSPSDBYKPPY-RSKUXYSASA-N
Application
- Schizophrenic behavior of 2,3-Oxidosqualene Sterol Cyclase from pig liver towards 2,3-oxidosqualene analogues. Alain Krief and colleagues discuss the variable catalytic behaviors of oxidosqualene cyclase when interacting with different oxidosqualene analogues, suggesting implications for sterol production efficiency and specificity (Krief et al., 2021).
Biochem/physiol Actions
Storage Class
10 - Combustible liquids
wgk_germany
WGK 3
flash_point_f
Not applicable
flash_point_c
Not applicable
Certificados de análisis (COA)
Busque Certificados de análisis (COA) introduciendo el número de lote del producto. Los números de lote se encuentran en la etiqueta del producto después de las palabras «Lot» o «Batch»
¿Ya tiene este producto?
Encuentre la documentación para los productos que ha comprado recientemente en la Biblioteca de documentos.
Los clientes también vieron
Nuestro equipo de científicos tiene experiencia en todas las áreas de investigación: Ciencias de la vida, Ciencia de los materiales, Síntesis química, Cromatografía, Analítica y muchas otras.
Póngase en contacto con el Servicio técnico