Recommended Products
product name
HMT-3522 S1, 98102210
biological source
human breast
growth mode
Adherent
karyotype
45,XX,1p-,-6,8q+-,12p+,14q+,17q+
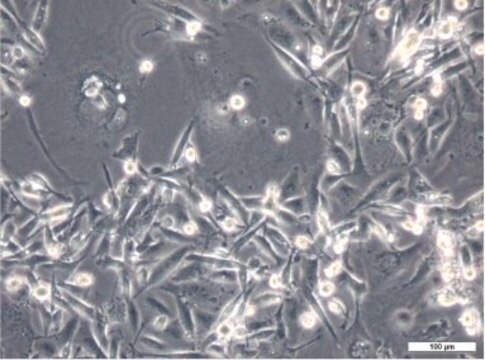
morphology
Epithelial-like
products
Not specified
receptors
EGF
technique(s)
cell culture | mammalian: suitable
relevant disease(s)
cancer
shipped in
dry ice
storage temp.
−196°C
Cell Line Origin
Human Caucasian breast epithelial
Cell Line Description
HMT-3522 S1, also known as HMT-3522/wt, is a subline that has been derived from HMT-3522. The parent line was established from a benign breast tumour of a 48 year old woman and has undergone spontaneous malignant transformation. It was grown on collagen in serum-free media without antibiotics from explantation onwards. A mutation at codon 179 of the p53 gene was detected during establishment of the parent line. HMT-3522 S1 is a continuation of this line from passage 34 growing without collagen. Subline S2 (Sigma catalog no. 98102211) has been isolated by culturing S1 cells from passage 118 in serum-free media without epidermal growth factor (EGF). The third subline, T4-2 (Sigma catalog no. 98102212) was established from S2 at passage 238 and is the only tumorigenic line in nude mice out of the 3 sublines. S2 and T4-2 grow on collagen-coated flasks. HMT-3522 S1 is near-diploid, but an evolution of karyotype has been observed by regular chromosomal analyses. The expression of cytokeratin 13, 14, 17 and 18 and vimentin has been reported. Expression of EGF-R, c-erb-B2, N-myc, c-ras-Ha1, c-ras-Ki2 and N-ras was shown to fluctuate slightly with passage number. The cells have tested negative for estrogen receptor, c-erbA1, c-erb-A2, int-2 or hst-1. During propagation growth factor requirements decreased simultaneously with amplification and overexpression of c-myc protooncogene. In a three-dimensional basement membrane culture assay the phenotypic characteristics of normal breast tissue in vivo are recapitulated. This cell line has remained non-tumorigenic for more than 400 passages. Publications using this cell line must refer to the original paper. Any sublines created must include the name HMT-3522 in the designation, but a short name may be used in the same paper.
DNA Profile
Not specified
Culture Medium
DMEM / Ham′s F12 (1:1) + 2 mM Glutamine + 250 ng/ml insulin + 10 μg/ml transferrin +10E-8M sodium selenite + 10E-10M 17 beta-estradiol + 0.5 μg/ml hydrocortisone + 5 μg/ml ovine prolactin + 10 ng/ml EGF
Subculture Routine
If starting from a frozen ampoule the cryoprotectant should be removed. Add thawed cells to a conical based centrifuge tube e.g. 15 ml tube; slowly add 4 ml of culture medium to the tube. Take a sample of the cell suspension, e.g. 100 ul, to count cells. Centrifuge the cell suspension at low speed, i.e., 100-150 x g for a maximum of 5 minutes.Split subconfluent cultures using 0.25% trypsin or trypsin/EDTA; 5% CO2; 37 °C. Seed pre-coated flasks at 2-4 x 10, 000 cells/cm2. Cells detach slowly; add same volume of trypsin inhibitor as trypsin used, centrifuge and reseed cells in collagen IV coated flasks. Medium is serum-free, therefore trypsin inhibitor is essential. S1cells can be slow in spreading out after subculture and may take 2-3 days to attach. Good growth occurs 5-7 days after subculture.
Cells should be frozen in (45% conditioned media: 45% fresh media) with 10% DMSO.
Cells should be frozen in (45% conditioned media: 45% fresh media) with 10% DMSO.
Other Notes
Additional freight & handling charges may be applicable for Asia-Pacific shipments. Please check with your local Customer Service representative for more information.
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service