Glycosaminoglycan Sulfation and Signaling
Vicki Caligur
Glycosaminoglycans are large linear polysaccharides constructed of repeating disaccharide units. The disaccharide units that are the basis of all glycosaminoglycans (except keratan) are a combination of an uronic acid (either glucuronic acid or iduronic acid) and an amino sugar (either N-acetyl-D-glucosamine or N-acetyl-D-galactosamine). There are four classifications of glycosaminoglycans:
- Hyaluronan hondroitin sulfate/dermatan sulfate
- eparan sulfate/heparin, and
- eratan sulfate.
A given glycosaminoglycan will contain only one type of amino sugar, but may contain both uronic acid forms, as iduronic acid is produced by enzyme-induced epimerization of glucuronic acid. The core disaccharide unit of keratan is N-acetyl-D-lactosamine ([1→3]β-D-galactose-β[1→4]-N-acetyl-D-glucosamine; β-D-Gal-[1→4]GlclNAc-[1→3]) and the galactose residue is not available for epimerization.
Glycosaminoglycan structures have a high degree of heterogeneity (termed “fine structure”) with respect to molecular weight, disaccharide construction, and sulfation. This variability can be attributed to the fact that glycosaminoglycan synthesis is dynamic. While protein and nucleic acid syntheses are template-based processes, glycosaminoglycan synthesis has no template but is modulated by the processing enzymes present. In addition, glycosaminoglycans, with the exception of hyaluronan, may have sulfate groups added to specific locations of the disaccharide subunit. However, enzymatic sulfation and epimerization reactions do not go to completion, resulting in structural heterogeneity within the polysaccharide molecule. This heterogeneity and dynamic processing make investigation of the structure/function relationships of glycosaminoglycans challenging.
Proteoglycans are proteins that have one or more glycosaminoglycan polymers attached to the protein core via an O-linkage through a serine residue. Glycosaminoglycans, whether separately or as a component of a proteoglycan, can bind to and interact with proteins. These glycosaminoglycan-protein interactions have essential roles in cellular processes including proliferation, cellular differentiation and development, and there have been efforts to relate specific sulfation patterns with resultant processes.
Hyaluronan is the only glycosaminoglycan that is not sulfated and does not attach to proteins to form proteoglycans. It has been associated with CD44 binding and signaling. The key function of keratan sulfate is as a structural component of cornea proteoglycans and not signaling effects. The glycosaminoglycans primarily involved in cellular signaling are heparan sulfate (HS)/heparin and chondroitin sulfate (CS)/dermatan sulfate (DS). This discussion will focus on heparan/heparin and chondroitin/dermatan and how sulfation impacts protein binding interactions and signaling effects by those glycosaminoglycans.
Biosynthesis of Heparan/Heparin and Chondroitin/Dermatan
The synthesis of heparan and chondroitin chains on a proteoglycan begins in the same way via an O-linked tetrasaccharide. The GAG chain synthesis is initiated by the attachment of xylose (Xyl) by xylosyltransferase (XylT) to a serine residue of the protein core. Two galactose (Gal) sugars are attached to the xylose sequentially by the enzymes galactosyltransferase I (GalT-I) and galactosyltransferase II (GalT-II). Glucuronyltransferase I (GlcAT-I) attaches the fourth sugar, glucuronic acid (GlcA) to the initial chain. At this point, the synthesis diverges (see Figure 1). A heparan polysaccharide continues with the attachment of an N-acetyl-D-glucosamine (GlcNAc) residue by N-acetylglucosaminyltransferase I. Alternatively, a chondroitin polysaccharide continues with the attachment of an N-acetyl-D-galactosamine (GalNAc) residue by N-acetylgalactosaminyltransferase I.1,2
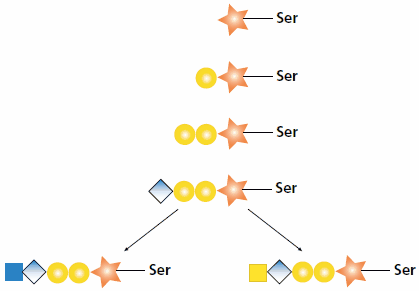
Figure 1.Biosynthesis of heparan and chondroitin chains. After the core tetrasaccharide (Xyl-Gal-Gal-GlcA-) is synthesized, the polymer can become a heparan chain by the addition of GlcNAc (shown left) or a chondroitin chain by the addition of GalNAc (shown right).
For heparan, the glycosaminoglycan is polymerized by the alternating addition of glucuronic acid and N-acetyl-D-glucosamine residues, to a final length of up to 200 disaccharides. The critical glycosyltransferases for heparan polymerization are exostosin 1 (EXT1) and exostosin 2 (EXT2). Modification of the polysaccharide is necessary for development of the heterogeneous fine structure. Heparan is modified by the N-deacetylase/N-sulfotransferase enzymes NDST1 and NDST2, which replace the N-acetyl groups (GlcNAc) with N-sulfate groups (GlcNS) on a glucosamine residue. The glucuronic acid units of the heparan chain are also susceptible to modification. Glucuronic acid (GlcA) may be converted to its epimer iduronic acid by the enzyme glucuronyl C5-epimerase. The iduronic acid moiety (IdoA) may be further modified by sulfation at the 2-O-position (IdoA-2S) through the activity of the enzyme heparan sulfate 2-O-sulfotransferase (HS2ST).2,3
These disaccharides may also be sulfated at the C-6 position of glucosamine (see Figure 2). Three variants of the heparan sulfate 6-O-sulfotransferase gene (HS6ST) have been identified.4 The uncommon form of sulfation is at the 3-O-position of the glucosamine ring and is mediated by the enzyme heparan sulfate 3-O-sulfotransferase (HS3ST). There are six isoforms of the HS3ST enzyme (1, 2, 3A, 3B, 4, and 5). Both the HS6ST and HS3ST enzymes have specific substrate preferences.
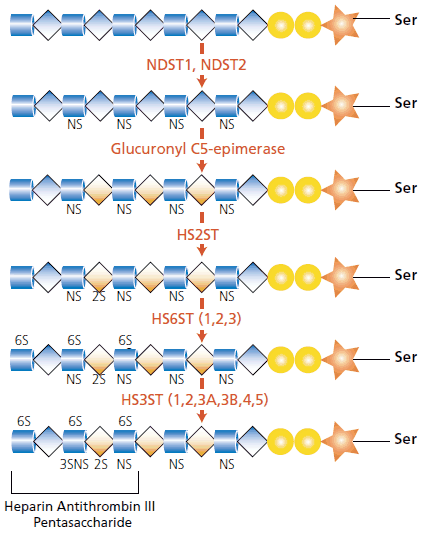
Figure 2.The modification of the heparan core is caused by a cascade of enzymes. The resultant heparan sulfate contains IdoA residues and domains with a high degree of sulfation. The pentasaccharide antithrombin II sequence associated with heparin is shown in the final image. See text for further explanation.
Sulfation is concentrated in specific regions of the heparan polysaccharide. As a result, heparan sulfate (HS) has regions of low sulfation interspersed with regions of high sulfation, with transitional regions separating the two. Highly sulfated domains, termed “composite sulfated regions” or S-domains, contain between 2 and 7 or 8 sequential GlcNS-IdoA-2S disaccharides. Other regions of the heparan polymer are largely unmodified and remain acetylated, constructed of GlcNAc-GlcA disaccharide units. Transition zones connect the two domains (see Figure 3). The fine structure complexity of HS is caused by both the variation in the disaccharide units and the overall variability created by the domain structure.5,6
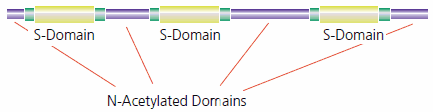
Figure 3.Heparan sulfate has regions of high sulfation (S-domains) interspersed with less modified regions (N-acetylated domains). A transition zone connects the two regions.
Heparin is a specific member of the heparan sulfate family of glycosaminoglycans. Heparin is expressed exclusively by mast cells, including mucosa mast cells, and heparin isolated from porcine intestinal cells has been approved by medical regulatory agencies for use as an anticoagulant. This anticoagulant activity of heparin has been shown to be due to the specific pentasaccharide (five-sugar) structure (see Figure 4).
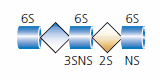
Figure 4.The pentasaccharide antithrombin binding site of heparin. Sites of sulfation are indicated.
This sequence contains a rare 3-O-sulfated glucosamine and is a specific ligand for the protease inhibitor antithrombin. Heparin binding of antithrombin blocks the activation of factors of the coagulation cascade mechanism. While heparin is frequently referred to in conjunction with heparan sulfate, it should be considered as a unique member of the heparan sulfate family and not interchangeable with heparan sulfate.2,7
For chondroitin synthesis, the glycosaminoglycan is polymerized by the alternating addition of glucuronic acid and N-acetyl-D-galactosamine residues. The completed polymer typically has 40 to over 100 disaccharide units. Modification of the chondroitin structure is also dynamic, generating fine structures that have type classifications that have not been applied to heparan. Chondroitins have been classified into 4 major types, based on location of sulfation on the disaccharide:
- Chondroitin sulfate A (CS-A) GlcA-GalNAc-4-SO4 (ΔDi-4S; A)
- Chondroitin sulfate C (CS-C) GlcA-GalNAc-6-SO4 (ΔDi-6S; C)
- Chondroitin sulfate D (CS-D) GlcA-2-SO4-GalNAc-6-SO4 (ΔDi-diSD; D)
- Chondroitin sulfate E (CS-E) GlcA-GalNAc-4,6-diSO4 (ΔDi-diSE; E)
Less common chondroitin disaccharides B, H, and K have also been identified.
Dermatan, also known as chondroitin sulfate B, undergoes significant epimerization of glucuronic acid (GlcA) to iduronic acid (IdoA). The enzyme 2-sulfotransferase preferentially sulfates IdoA adjacent to GalNAc-4-SO4. As a result of this enzymatic specificity, the primary disaccharide unit of dermatan sulfate is IdoA-2-SO4-GalNAc-4-SO4 (iB). Other rare dermatan sulfates have been recognized, and are named similarly to their chondroitin sulfate counterparts (iA, iC, iD, and iE), using the letter “i” to indicate the iduronic acid residue.
As the sulfation and epimerization reactions occur to varying degrees, there is no absolute structure of any chondroitin sulfate/dermatan sulfate, and chondroitin/dermatan contains many fine structures much like heparan sulfate.1
Heparan Sulfate Signaling
Heparan sulfate proteoglycans (HSPGs) have been extensively studied for their role in extracellular signaling. HSPGs interact with and regulate growth factors, chemokines, and cytokines. Signaling pathways for apoptosis, cellular development, and adhesion are regulated by proteoglycans. Genetic defects in heparan synthesis and modifying enzymes may affect growth factor signaling and produce morphological defects.8 Drosophila mutants and gene knockout mouse models have been used to eliminate critical heparan synthesis and modification enzymes, and subsequently identify the developmental processes disrupted. Experiments that use Drosophila containing mutations of the protein core of proteoglycans have been performed and have shown that the protein core also affects development. It has been theorized that both the HS and the protein core have roles in the mediation of growth factor binding and cell signaling.6
HS and HSPGs participate in fibroblast growth factor-2 (FGF-2) activation in response to injury and inflammation. After injury, HSs break free from the cell membrane or their proteoglycan serine linkage and become soluble. Heparanase that is released by cells involved in inflammation cleaves the released heparan sulfate into smaller heparin-like oligosaccharides. While intact HS does not activate FGF-2, these smaller oligosaccharides are able to activate FGF-2. This processing of free HS by heparanase in response to inflammation has been theorized to be a necessary step to wound healing.2,9
The binding of HSPGs to FGF-2 has been studied as well for a possible association with malignant transformation of breast carcinoma cells by FGF-2 in vitro. Fibroblast growth factors require the presence of heparan sulfate or heparin to stabilize binding with tyrosine kinase signaling receptors (FGFR-1 through FGFR-4). Mundhenke found that HSPGs isolated from the MCF-7 breast-carcinoma cell line had an elevated ability to promote formation of the FGF-2/HSPG/FGFR-1 complex as compared to normal epithelial cells. HSPGs have also been shown to affect signaling by attachment to other growth factors including FGF-7.10
Using glycosaminoglycan microarrays, Shipp and Hsieh-Wilson showed that individual members of the FGF family require specific sulfation patterns for HS binding. In addition to confirming sulfation specificities previously reported for FGF-1, FGF-2, FGF-4, they were able to characterize the heparan sulfation requirements for FGF-16 and FGF-17. Using the microarrays, chemotactic proteins, including slit2, that regulate axon extension were found to have preferential binding to specific sulfation patterns. This finding was validated by co-culturing embryonic olfactory bulb cells with HEK293 cells expressing slit2. The cultures were supplemented with heparan sulfate variants containing different sulfation patterns. After staining with anti-τ antibody for axon visualization, the cultures showed that highly sulfated heparan sulfate reduced slit2-mediated axon repulsion, while desulfated heparin or N-acetylated heparin had no effect on slit2 repulsion.11
The genes for the heparan polymerases exostosin 1 (EXT1) and exostosin 2 (EXT2) were initially thought to be tumor suppressor genes. Mutations of EXT-type genes in Drosophila reduced the level of heparan synthesis and inhibit hedgehog (Hh), Wingless (Wg, which correlates to Wnt), and Dpp signaling pathways, indicating that HS participates in the signaling function of these pathways.12,13 The protein core of the proteoglycan has been shown to contribute to HSPG signaling effects as well. Wnt signaling was modulated by the processed protein core of the HSPG glypican 3, with the implication that proper signaling requires contributions by both the HS and the core protein components.14
A similar protein-glycosaminoglycan synergy toward binding affinity was identified for the association of interleukin-8 (IL-8) with the HSPG syndecan 2 on human umbilical vein endothelial cells. IL-8 is a chemokine that is secreted at sites of inflammation by cytokine activated endothelial cells and which is retained on the cell surface by interactions with HSPGs. While glycosylated syndecan 2 showed strong affinity for IL-8, the nonglycosylated protein core also demonstrated weak IL-8 binding.15
CD44 (Pgp-1, ECM-III, H-CAM) functions as a homing receptor and is most commonly the cellular receptor for hyaluronic acid. Jones, et al., found that some CD44 isoforms exist as heparan sulfate proteoglycans, and that CD44/HSPG was able to bind FGF-2, vascular endothelial growth factor (VEGF), and heparinbinding epidermal growth factor. CD44/HSPG isoforms were overexpressed by macrophages within inflamed rheumatoid arthritis tissue but minimally expressed in noninflamed osteoarthritis tissue. Based on the experimental data, it was theorized that CD44/HSPG isoforms are involved in the regulation of growth factor activity in inflammatory arthritis.16
You, et al., reported that the tumor necrosis factor receptor family member Decoy receptor 3 (DcR3) could induce apoptosis in dendritic cells (DC) through binding between DcR3 and DC surface heparan sulfate proteoglycans This was based on the observation that HBD.Fc, the recombinant protein containing both the heparan sulfate-binding domain (HBD) of DcR3 and the Fc portion of human IgG1, functions in the same manner as DcR3.Fc to induce apoptosis in dendritic cells.17
In addition to sulfate modifications present on the heparan chain, cell surface sulfatases are critical to growth factor signaling. The human sulfatases Sulf1 and Sulf2 are 6-O-endosulfatases and are able to remove 6-O-sulfate from the highly sulfated regions of the polysaccharide.6 The association between 6-O-sulfation of HS and FGF signaling has been studied from many aspects. In a review by Nakato and Kimata, the authors discussed how the fine structures of HS, especially those containing 6-O-sulfation, modulate the growth factor signaling functions of HSPGs.8 Increased Sulf-1 expression by the breast cancer cell line MDA-MB-468 inhibited angiogenesis and cancer cell proliferation in vivo. The inhibition was attributed in part to the reduced ability of the vascular HS to form a stable FGF-2/HSPG/FGFR-1 complex. The inference is that Sulf-1 overexpression reduces the degree of 6-O-sulfation in HS fragments, and that 6-O-sulfation contributes to the stabilization of the FGF-2/HSPG/FGFR-1 complex.18 More recently, experiments with murine embryonic fibroblasts (MEF) HS-6-O-transferase deficient mice (HS6ST-1-KO, HS6ST-2-KO, and double knock-out) showed that FGF-2 and FGF-4-dependent signaling in MEF was significantly reduced compared to that of wild-type fibroblasts.19
The vascular endothelial growth factor VEGF165 contains a heparinbinding domain that is only weakly activated by 2-O-desulfated heparin or 6-O-desulfated heparin.20 6-O-sulfation has also been shown to affect the signaling pathways of Wnt and bone morphogenic protein (BMP). It has been suggested that there is a dynamic interaction between the Sulf enzymes and HS biosynthetic enzymes, rather than independent processes.6
Chondroitin Sulfate/Dermatan Sulfate Signaling
While HS has been of primary interest in glycosaminoglycan signaling effects, chondroitin sulfate (CS) and dermatan sulfate (DS) have also been recognized as affecting signaling pathways,21 and that these interactions are dependent on the polysaccharide fine structure. Both the glycosaminoglycan components and the proteoglycan have been investigated for contributions to biological function. Because of the fine structure and heterogenetity, both CS and DS units may exist as hybrids.22 Research has primarily focused on the contribution by the specific chondroitin suflate proteoglycans (CSPGs) and dermatan sulfate proteoglycans (DSPGs) aggrecan, decorin, biglycan, versican and the syndecans. Additional CSPGs/DSPGs of interest are neurocan, brevican, bamacan, betaglycan, serglycin, endocan, and appican.
In much the same manner as HS, CS/DS and CSPGs /DSPGs participate in fibroblast growth factor activation in response to injury and inflammation.2,22 After injury, the glycosaminoglycan chains are cleaved at their serine linkage, or the proteoglycans separate from the cell membrane and become soluble. Soluble dermatan sulfate is able to activate FGF-2 23 and FGF-7, initiating cellular proliferation via FGF-7 through the mitogen-activated protein-kinase (MAPK) pathway.24 The minimum dermatan sulfate structure required by FGF-2 is an octasaccharide and the requirement by FGF-7 is a decasaccharide. 4-O-Sulfation is also required for FGF-dependent cell proliferation, while additional 2-O-sulfation or 6-O-sulfation did not increase the disaccharide activity.25
The hepatic growth factor/scatter factor (HGF/SF) is known to participate in morphogenesis and cellular differentiation, organogenesis, and angiogenesis. Endocan, a DSPG that is regulated by proinflammatory cytokines, requires the presence of the glycosaminoglycan moiety in order to promote HGF/SF-induced proliferation of human embryonic kidney cells.26 HGF/SF binding demonstrated preferential binding by 2,6-O-disulfated DS and highly sulfated HS, but not CS, indicating a selective requirement for either iduronic acid residues or a high degree of sulfation, although highly sulfated polysaccharides were less bound than the iduronic acid-containing glycosaminoglycans.27
While the fine structure of CS/DS affects binding and signaling effects, it turns out that growth factors can modulate the expression and structure of the CS/DS glycosaminoglycan. Cultured human lung fibroblasts were treated with transforming growth factor-β1 (TGF-β1) alone, or in combination with epidermal growth factor and platelet-derived growth factor. Cells treated with TGF-β1 or the combination of growth factors increased their expression of CS/DS proteoglycans. In addition, the sulfation and epimerization profiles of the chondroitin/dermatan chains were significantly modified compared to untreated fibroblasts.28
The highly sulfated CS-E type has been shown to bind to heparin binding growth factors midkine (MK), pleiotropin (PTN), heparin-binding epidermal growth factor-like growth factor (HB-EGF), FGF-16, and FGF-18. As many of these growth factors are expressed in the mammalian brain, it was proposed that CS-E and CSPGs are critical to the development of the brain and central nervous system.29 Subsequent work using an antibody specific for both the CS-E disaccharide and the oversulfated DS-E disaccharide (IdoA-GalNAc-4,6-diSO4; iE) revealed the presence of these glycosaminoglycan sequences in the mouse developing brain. The strong expression of the gene for GalNAc4S-6ST was associated with brain development, and demonstrated that E/iE-containing CS/DS chains are critical in brain development with the implication that E/iE-containing CS/DS chains participate in neurogenesis, axon guidance, and/or neuronal survival.30
While the glycosaminoglycan sequence and sulfation pattern regulate signaling, the protein core of the glycosaminoglycan also contributes to the signaling ability. The ligand binding of the CS/DS proteoglycans decorin and biglycan to transforming growth factor-β (TGF-β), tumor necrosis factor-β (TNF-β), and Wnt-induced secreted protein-1 (WISP1) is mediated by both the DS component and the protein cores. A recent review by Seidler and Dreier analyzed both the galactosaminoglycan component and the protein core of decorin as critical to the ability of decorin to function as a ligand for other signaling molecules.31
The enzymes N-acetylgalactosamine-4-sulfatase (arylsulfatase B; ASB) and galactose-6-sulfatase (GALNS) hydrolyze sulfate groups of CS/DS. MCS-7 cells that overexpressed ASB or GALNS produced increased expression of CS/DS proteoglycans syndecan-1 and decorin. Alernatively, when siRNA was used to silence ASB or GALNS expression in MCS-7 cells, the level of CS content increased and syndecan-1 expression was inhibited. The results demonstrate a role for these sulfatases in CS development and the regulation of proteoglycan expression.32
Research Techniques
A variety of experimental techniques have been employed to identify the structure of the glycosaminoglycan chain, the degree and location of sulfation, and the relationship between polysaccharide component and protein core. Knock out mice that lack a designated heparan or chondroitin/dermatan synthesis enzyme have been used to help identify the pathways affected by the lack of glycosaminoglycan modification. Table 1 lists several of the enzymes involved in glycosaminoglycan synthesis and modification and their genes. Table 2 lists the genes for proteoglycan core protein expression. RNA interference has been applied in vitro as another means to eliminate synthesis and modification enzymes to study downstream cellular processes. ELISA analysis with proteoglycan core specific antibodies has been used to monitor protein expression, while quantitative RT-PCR has been applied to study gene regulation.
Genes of Heparan and Chondroitin Biosynthesis Enzymes |
|---|
Genes of Proteoglycan Core Proteins |
|---|
Enzymatic degradation is often used to cleave glycosaminoglycans into disaccharide units for subsequent analysis. Amide-hydrophilic interaction chromatography (HILIC) liquid chromatography with tandem mass spectrometry (LC/MS/MS) has been applied for comparative glycomics of chondroitin sulfate isoforms. As carbohydrates are inherently difficult to detect by HPLC, they are often modified by reductive amination to covalently attach a detection molecule, typically 2-aminobenzoic acid (2-AB), 2-anthranilic acid (2-AA) or 2-aminopyridine (2-AP). Stable isotopically labeled 2-anthranilic acid was used to comparatively profile CS disaccharides from different tissues.33
Poly-L-lysine coated slides were used to create microarrays of desulfated HS glycosaminoglycans. The microarrays were used to profile the binding of FGF-2 and other FGF factors against different sulfation patterns. The microarray technique provides a high throughput method to study protein-glycosaminoglycan binding.11
High-resolution NMR spectroscopy, X-ray crystallography, and molecular modeling are valuable methods applied to the investigation of glycosaminoglycan-protein structural interactions. Some of the resulting glycan-protein data has been recorded and is publicly available from the Centre de Recherches sur les Macromolécules Végétales (CER-MAV; http://www.cermav.cnrs.fr/glyco3d).34
A glycomics approach has been used to monitor the regulation of glycosaminoglycan biosynthesis during differentiation of mouse embryonic stem cells. Glycosaminoglycan content and composition changed during cellular transition from stem cells to embryoid bodies and extraembryonic endodermal cells, with 4 to 6-fold increases in CS/DS synthesis and 5 to 8-fold increases in heparan sulfate synthesis. mRNA transcription levels for glycosaminoglycan synthesis and modifying enzymes were unchanged and did not appear to correlate to the changes in glycosaminoglycan levels found during differentiation.35
Summary
Much has been learned in the last few years about glycosaminoglycans. The synthesis mechanisms have been elucidated in great detail and their contribution to proteoglycan structure recognized. The relationship of fine structure and core protein structure to receptor binding is now known to be necessary for multiple cellular processes. Techniques used for genomics, proteomics, and cell biology have been successfully applied to the investigation of glycosaminoglycan signaling pathways. While some aspects of the structural requirements of glycosaminoglycans and proteoglycans for effective signaling have been identified, the reasons for why they participate in cellular processes and how the signaling pathways are initiated and regulated are still to be determined.
References
Para seguir leyendo, inicie sesión o cree una cuenta.
¿No tiene una cuenta?