DNA Oligonucleotide Synthesis
Phosphoramidite chemistry, developed in the 1980s and later enhanced with solid-phase supports and automation, is the method of choice for DNA oligonucleotide manufacturing. Opposite of biosynthesis, chemical synthesis proceeds in the 3' → 5' direction according to the steps outlined in Figure 1. Note: this article focuses exclusively on DNA synthesis (while similar, there are differences with the chemical synthesis of RNA).
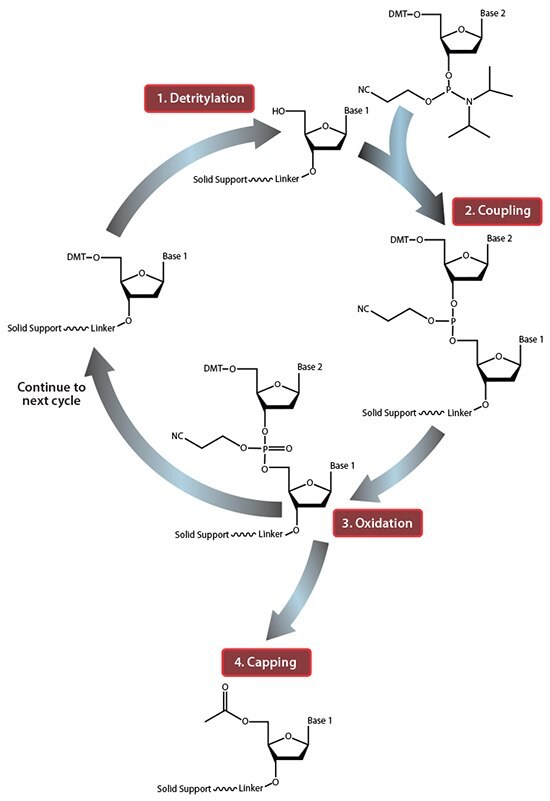
Figure 1.Summary of solid-phase oligonucleotide synthesis cycle. In step 1, Detritylation, the 5'-DMT protecting group is removed from the first, solid-support-linked nucleoside. In step 2, Coupling, the free 5’-OH of the first, solid-support-linked nucleoside attacks the phosphorus of the incoming second nucleoside, displacing its diisopropylamino group. In step 3, Oxidation, the unstable phosphite triester is converted to a stable phosphate triester, which allows the next cycle to proceed to step 1, Detritylation of the second nucleotide. However, prior to moving to the next cycle, in step 4, Capping, solid-support-linked nucleosides with unreacted 5’-OH are acetylated, thereby preventing elongation of sequences with deletion mutations (Capping is performed after Oxidation to drive all water out, which would otherwise inhibit the next cycle of the reaction).
Oligonucleotide Synthesis Phosphoramidite Method
This section explores the detailed chemistry of each of the four steps [step 1 (detritylation), step 2 (coupling), step 3 (capping), and step 4 (oxidation)] in the phosphoramidite method.
Step 1 (Detritylation)
The cycle is initiated by removal of the 5'-DMT (4,4'-dimethoxytrityl) protecting group of the solid-support-linked nucleoside (contains the terminal 3' base of the oligonucleotide). The 5'-DMT prevents polymerization of the nucleoside during functionalization of the solid support resin. The mechanism is displayed in Figure 2.

Figure 2.The detritylation mechanism. The 5'-DMT protecting group is removed by TCA (trichloroacetic acid) in the solvent dichloromethane (too concentrated a solution of TCA or too long of a detritylation time leads to depurination and hence, lowers the overall yield of the final oligonucleotide). The products include the 3' terminal nucleoside with a free 5'-OH and a DMT carbocation (resonance structure formed by electron delocalization not shown). The nucleoside proceeds to step 2 in the synthesis while the DMT carbocation absorbs at 495 nm and thereby produces an orange color that can be used to monitor coupling efficiency.
Step 2 (Coupling)
Once the DMT has been removed, the free 5'-OH of the solid-support-linked nucleoside is able to react with the next nucleoside, which is added as a phosphoramidite monomer. The mechanism is displayed in Figure 3.
![The coupling mechanism. The diisopropylamino group of the incoming phosphoramidite monomer in the solvent acetonitrile is ‘activated’ (protonated) by the acidic catalyst ETT [5-(ethylthio)-1H-tetrazole]. The mixing is carried out in the fluid lines of the synthesis instrument as the reagents are delivered to the solid support. The activated phosphoramidite is delivered in a many-fold excess over the solid-support-linked nucleoside to drive the reaction to as close to completion as possible. The products include a dinucleoside with a phosphite triester linkage and a free diisopropylamino group. The coupling mechanism](/deepweb/assets/sigmaaldrich/marketing/global/images/technical-documents/articles/genomics/pcr/dna-figure-3/dna-figure-3.jpg)
Figure 3.The coupling mechanism. The diisopropylamino group of the incoming phosphoramidite monomer in the solvent acetonitrile is ‘activated’ (protonated) by the acidic catalyst ETT [5-(ethylthio)-1H-tetrazole]. The mixing is carried out in the fluid lines of the synthesis instrument as the reagents are delivered to the solid support. The activated phosphoramidite is delivered in a many-fold excess over the solid-support-linked nucleoside to drive the reaction to as close to completion as possible. The products include a dinucleoside with a phosphite triester linkage and a free diisopropylamino group.
Step 3 (Oxidation)
The phosphite triester formed during the coupling reaction is unnatural and unstable; therefore, it must be converted to a more stable phosphorus species prior to the start of the next cycle. Oxidation converts the phosphite triester to the stable phosphate triester. The mechanism is displayed in Figure 4.

Figure 4.The oxidation mechanism. Oxidation of the phosphite triester is achieved with iodine in the presence of water and pyridine. The product is the phosphate triester, which is essentially a standard DNA backbone with a β-cyanoethyl protecting group on the free oxygen.
Step 4 (Capping)
Since 100% coupling efficiency is impossible, there are always some solid-support-linked nucleosides with unreacted 5'-OH. If not blocked, these hydroxyl groups will react during the next cycle, and hence, lead to a missing base. The accumulation of these deletion mutations through successive cycles would create a complex mixture of ‘shortmers’ that are difficult to purify and therefore could render the oligonucleotide useless in the subsequent application. Capping is required to prevent shortmer accumulation. The mechanism is displayed in Figure 5.

Figure 5.The capping mechanism. Acetic anhydride and N-methylimidazole react to form an intermediate in the solvent tetrahydrofuran, which contains a small quantity of pyridine. The mixing is carried out in the fluid lines of the synthesis instrument as the reagents are delivered to the solid support. The product is the solid-support-linked nucleoside with an acetylated 5'-OH (pyridine maintains a basic pH thereby preventing detritylation of the phosphoramidite monomer by the free acetate / acetic acid).
Successive Cycles
The second cycle begins by starting with step 1, detritylation, followed by each of the remaining three steps. The number of cycles repeated equals the desired number of bases. We synthesize oligonucleotides from 2 to 120 bases.
Cleavage
Our proprietary solid support / linker is stable to all phosphoramidite reagents but is cleavable from the oligonucleotide at the end of synthesis. Cleavage is necessary so that the free 3'-OH may take part in biochemical reactions, such as extension by DNA Polymerase during PCR when the oligonucleotide serves as a primer. The reactant and product are displayed in Figure 6.

Figure 6.The reactant and product of cleavage. Ester hydrolysis of the linker (and, simultaneous removal of the solid support) is carried out by treatment with concentrated aqueous ammonia. The product is the oligonucleotide with a terminal, free 3'-OH.
Deprotection
After cleavage, the solution of oligonucleotide in concentrated aqueous ammonia is heated to remove protecting groups from the bases and phosphates.
Bases
While thymine does not require a protecting group, adenine, cytosine, and guanine do since they contain exocyclic primary amino groups. The protecting groups must be removed so that proper hydrogen bonds between the oligonucleotide and the target nucleic acid may form. The reactant and product are displayed in Figure 7.

Figure 7. The reactant and product of base deprotection. The oligonucleotide in concentrated aqueous ammonia is heated. The protecting groups include: N(6)-benzoyl A, N(4)-benzoyl C, and N(2)-isobutyryl G. The product of the reaction is fully-deprotected A, C, and G bases.
In addition to the standard protecting groups, the labile dimethylformamidyl G and the ‘ultramild’ protecting groups can be used for modified oligonucleotides that are sensitive to ammonia. These protecting groups on the relevant bases are displayed in Figure 8.

Figure 8.Labile and ultramild protecting groups. The dimethylformamidyl protecting group is typically removed in concentrated aqueous ammonia via heating but in significantly less time than is the isbutyryl group. The ultramild protecting groups include: N(6)-phenoxyacetyl A, N(2)-acetyl C, and N(2)-isopropylphenoxyacetyl G. They are typically removed at room temperature in a concentrated aqueous ammonia / methylamine solution.
Phosphodiester Formation
The β-cyanoethyl group on the free oxygen of the phosphate must be removed to convert it from a phosphate triester to a phosphate diester (phosphodiester). The mechanism is displayed in Figure 9.

Figure 9.The mechanism of phosphodiester formation via deprotection. The cyanoethyl groups are removed in concentrated aqueous ammonia via β-elimination. The reaction is quick because the hydrogen atoms on the carbon adjacent to the electron-withdrawing cyano group are highly acidic. The products are the oligonucleotide with a native phosphodiester backbone and acrylonitrile.
Yield
The yield of the synthesis is heavily dependent on the coupling efficiency. Even with 99.5% efficiency, which we readily achieve, you can see in Table 1 that as the oligonucleotide length increases, the yield rapidly decreases.
It is important to keep in mind that coupling efficiency is not all that determines the yield. Deprotection, cleavage, and purification (even simple desalting) decrease the yield further. Our guaranteed yields based on manufacturing scale and purification can be found here.
Para seguir leyendo, inicie sesión o cree una cuenta.
¿No tiene una cuenta?