DNA Standards for Metagenomic Research into Microbiome Communities
Microbiome research has contributed valuable data that provides critical insight into the nature of microbial communities. As research in the microbiome field evolves, it has become evident to the scientific community that basic tools for internal standardization and inter-sample normalization are essential for the microbiome metagenomic workflow. The most effective way to ensure data integrity is by using standards.
- THE FUNCTION OF STANDARDS IN MICROBIOME RESEARCH
- HOW ARE DNA STANDARDS USED IN MICROBIOME RESEARCH?
- DNA STANDARDS: ESSENTIALS FOR THE FUTURE OF MICROBIOME METAGENOMIC RESEARCH
- SELECTION OF THE APPROPRIATE DNA STANDARD FOR MICROBIOME ANALYSIS
- EXTREMOPHILE DNA AS CONTROLS, REFERENCE, AND NGS WORKFLOW STANDARDS
- FUNGAL AND VIRAL DNA STANDARDS IN MICROBIOME ANALYSIS
- SELECT THE RIGHT DNA STANDARDS FOR THE MICROBIAL COMMUNITY OF INTEREST
- DNA STANDARDS FOR METAGENOMIC ANALYSIS OF THE MICROBIOME
THE FUNCTION OF STANDARDS IN MICROBIOME RESEARCH
Standards are known concentrations of substances used in quantitative analysis. A standard provides a reference that can be used to determine concentrations or quantities of substances in unknown samples. Microbiome DNA standards enable
- Result validation and comparison of metagenomic workflows, both for specific microbes and mixed microbial communities
- Enhanced confidence in reliable, reproducible results
- More consistent assay protocols
- Elimination of biases and enhanced quality control
- Improved reproducibility and normalization among different labs
- Establishing a base for comprehensive microbial metadata collections
DNA standards are increasingly recognized for enabling reproducibility and normalization between studies across different research trials, thus helping to establish a base for comprehensive metadata collections.
HOW ARE DNA STANDARDS USED IN MICROBIOME RESEARCH?
Standardization can be utilized throughout the workflow for metagenomics analysis of the microbiome, from sample preparation and extraction to NGS sequencing, and bioinformatics analyses.2 Types of standards that enhance credibility and reproducibility in microbiome research include
- Microbial community DNA standard mixtures
- Extremophile DNA standards for spike-in
- Inactivated bacteria standards


DNA Extraction
- DNA-free reagents
- Inactivated bacteria standards

Library Prep
- Extremophile DNA spike-in standards
- Microbial community DNA standard mix
- Inactivated bacteria standards

DNA Sequencing
- Extremophile DNA spike-in standards
- Microbial community DNA standard mix
- Inactivated bacteria standards

Data Analysis
- NGS analysis
DNA STANDARDS: ESSENTIALS FOR THE FUTURE OF MICROBIOME METAGENOMIC RESEARCH
Standardization is appropriate throughout the metagenomics workflow, from common bench processes such as sample preparation and amplification to NGS sequencing and bioinformatics analyses. Without standardization, the validity of results and their meaningful comparison is compromised. To maintain unbiased research, there must be an indicator that can ensure appropriate comparison of results from different samples or donors, and from laboratory to laboratory.
The parameters of each human microbiome sample can be diverse with respect to donor age, gender, and health background. A lack of standardization can lead to biases and errors in common processes during sample preparation and analysis such as amplification, sequencing, and bioinformatics investigations. Without standards, there is no way to normalize results for subsequent research, which can impair the development of novel medical applications.
SELECTION OF THE APPROPRIATE DNA STANDARD FOR MICROBIOME ANALYSIS
We offer human-derived microbial DNA standards, environmental DNA standards and inactivated whole cell standards. Our microbial DNA standards for the microbiome NGS metagenomics workflow are offered as single-strain genomic DNA, or as a mixed community of genomic DNA to use as controls. These standards come in a range of genome sizes and GC content, and are available for analysis of diverse phyla. We offer DNA from environmentally-derived strains that can be used as internal qualitative and quantitative controls when spiked into a microbiome sample.
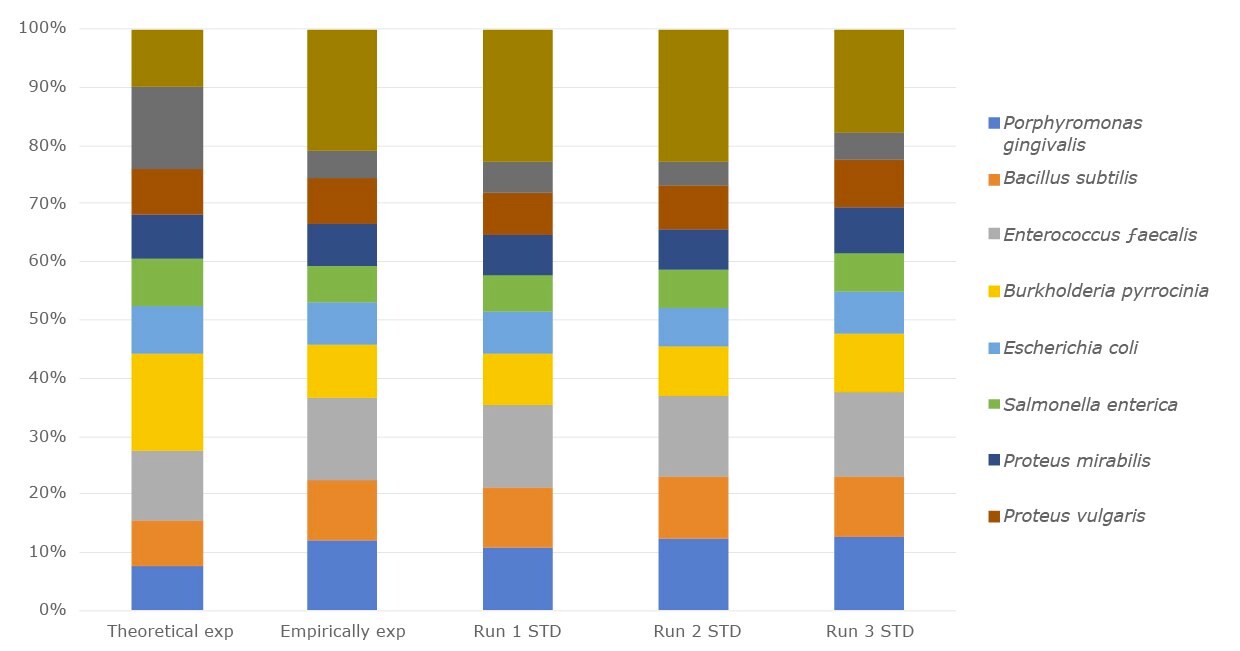
Figure 1.The relative abundance of the DNA microbial community mix from three separate experiments compared with empirical expected output and the theoretical expected output.
Inactivated whole cell standards for microbiome analysis
Inactivated whole cell standards are essentially dead strains - specific bacteria that can be included as a discrete sample, or as a spike-in during collection and extraction that serves as a quality control for assay optimization or a benchmarking control tool. These standards are an excellent control when added to the workflow in the initial stages and are ideal for evaluating the accuracy and reproducibility of a workflow from sample collection to metagenomic analyses.
DNA standards may be provided as single bacteria-specific strains, or as a communal mix to be added during preparation of the library. Standards may either be spiked in to the test samples, or included as a separate sample in the run as a comparative index for results from a workflow. The benefits of DNA standards are numerous: they provide a reliable way to check whether a sample is degraded or contaminated; they provide relative abundance comparisons; and they ensure the normalization of the workflow.
EXTREMOPHILE DNA AS CONTROLS, REFERENCE, AND NGS WORKFLOW STANDARDS
Extremophile standards (inactivated whole cell or DNA) can be used for benchmarking the performance of the workflow from the initial sample preparation to the meta-genomics analyses, and can be a tool for enhancing reproducibility and accurate comparison of workflow results. Because extremophile bacteria are unlikely to be found in the human microbiome, standards from these species can be used as internal standards for the NGS workflow, as a control for sample preparation, or as a reference DNA for checking sequencing accuracy.
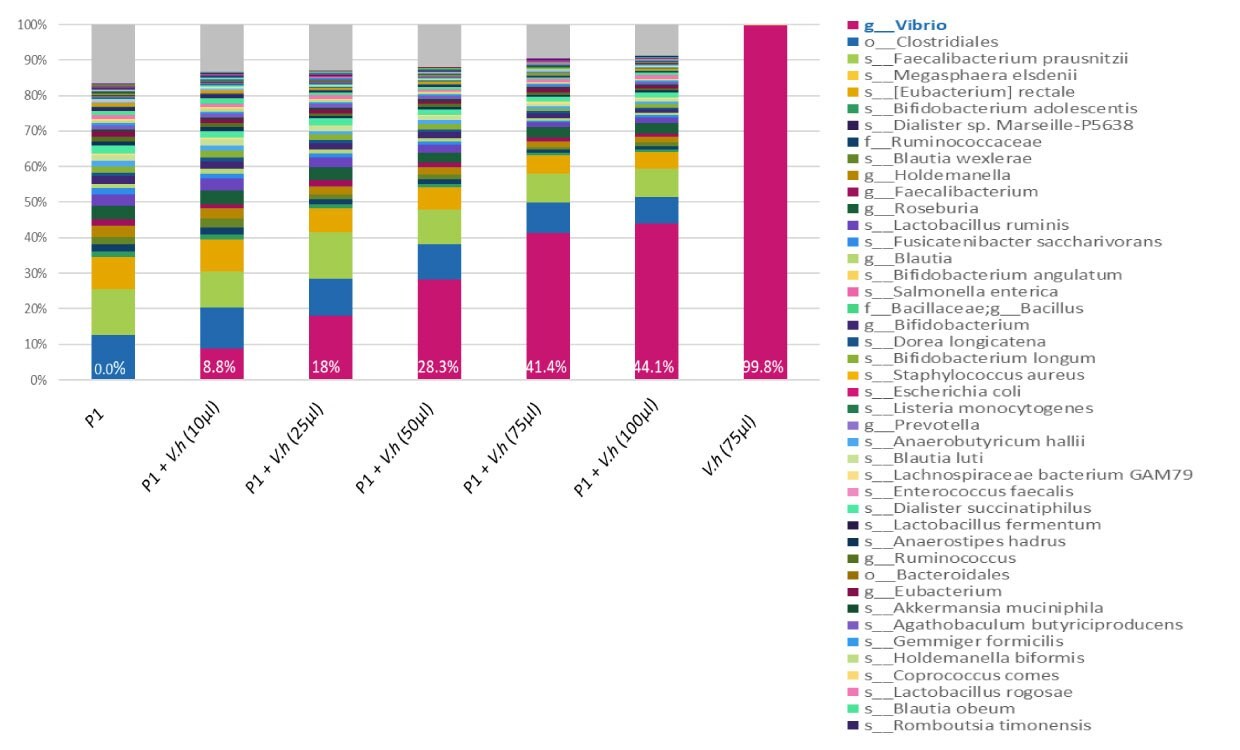
Figure 2.A range of concentrations (10, 25, 50, 75, and 100 µl) of inactivated bacteria V. Harveyi MBD0037 (pink) were spiked into the same human stool sample (P1). DNA was amplified with 16S V3-V4 primers and sequenced on Miseq (250 bp reads, paired-end). When MBD0037 (pink) is spiked into a human fecal sample it can be clearly identified by 16S NGS analysis. MBD0037 can therefore serve as an internal spike-in standard control in a human microbiome genomics workflow.
FUNGAL AND VIRAL DNA STANDARDS IN MICROBIOME ANALYSIS
The term ‘microbiome’ was first used in 1952 to describe all of the microorganisms in a specific environment. In addition to bacteria and archaea, it has become clear that the role of both fungi and viruses in the microbiome is increasingly relevant and can have a significant impact on human health. Specialized standards are therefore also used to help normalize and serve as a control for specific organisms in the microbiome such as fungi and viruses.
Our growing catalog of DNA standards includes those for Aspergillus fumigatus and Candida albicans, both of which are associated with the human fungal microbiome, also known as the human mycobiome.
Select the right DNA standards for the microbial community of interest
Microbial communities differ significantly in species composition. In the human body, for example, microbiome environments vary significantly by anatomical site (gut, skin, mouth, vagina)—so it is imperative that standards selected are relevant to the microbial diversity that is characteristic of the environment, whether from individual strains or community mix standards. Strains that are not part of the site-specific microbial composition can be spiked in to benchmark workflow steps and serve as controls. For example, using extremophiles will be beneficial when sampling diverse human microbial communities, since they are not found in the human body. By contrast, using a community mix of typical gut microbes in a skin microbial sample can cause errors and biases that will yield metagenomic results that can be easily misinterpreted, and that can impact the research.
Learn more about our complete microbiome research offering.
Inactivated Whole Cell Standards
Mixed Community Bacterial DNA Standards
Fungal (Mycobial) DNA Standards
References
Para seguir leyendo, inicie sesión o cree una cuenta.
¿No tiene una cuenta?