推薦產品
一般說明
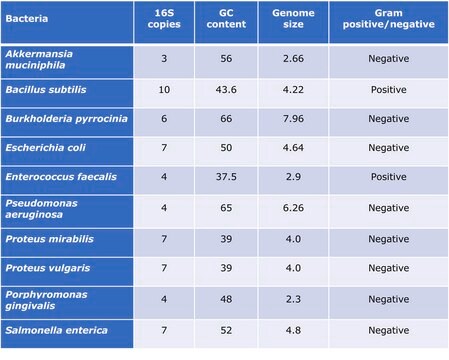
Standardization of sample analysis is essential in microbiome genomics research workflow. Lack of standardization can lead to biases and errors in common processes during sample preparation and analysis such as sample amplification, sequencing, and bioinformatics analyses.1 The human microbiome includes not only bacteria, but also viruses and fungi. While the “bacteriome” field has been researched extensively in the past years, fungi including yeasts, comprising the “mycobiome”, remain relatively neglected. Fungi and yeasts have been shown to be involved in illness conditions such as cancer and gastrointestinal disease. The human gut is populated by three fungal phyla, Ascomycota, Basidiomycota, and Zygomycota.
The fungal microbial genomic DNA standard from Candida albicans can serve as standard for benchmarking the performance along the workflow of microbiome research and metagenomic analyses, as well as a tool to increase reproducibility and allow comparison of results obtained by different labs.
Candida albicans is an opportunistic human fungal pathogen belonging to Ascomycota. It exists as a harmless commensal in the gastrointestinal and genitourinary tracts in about 70% of humans and about 75% of women suffer from Candida infection at least once in their lifetime. C. albicans has emerged as a model organism for studying fungal pathogens along with other two fungi: Aspergillus fumigatus and Cryptococcus neoformans.2
Read here how to use our standards to ensure data integrity for your microbiome research.
The fungal microbial genomic DNA standard from Candida albicans can serve as standard for benchmarking the performance along the workflow of microbiome research and metagenomic analyses, as well as a tool to increase reproducibility and allow comparison of results obtained by different labs.
Candida albicans is an opportunistic human fungal pathogen belonging to Ascomycota. It exists as a harmless commensal in the gastrointestinal and genitourinary tracts in about 70% of humans and about 75% of women suffer from Candida infection at least once in their lifetime. C. albicans has emerged as a model organism for studying fungal pathogens along with other two fungi: Aspergillus fumigatus and Cryptococcus neoformans.2
Read here how to use our standards to ensure data integrity for your microbiome research.
應用
Suitable as a standard for PCR, Sanger and next generation sequencing (NGS).
特點和優勢
- Individual microbial standard for microbiomics and meta-genomics workflow
- Suitable standard for PCR, Sanger sequencing and 18S NGS
- Improves bioinformatics analyses
- Increases reproducibility and repeatability
- Identity and purity of the genomic Candida albicans DNA has been confirmed by 18S NGS and gel electrophoresis
外觀
Liquid -The genomic DNA is provided at ≥10 ng/μl concentration in TE buffer pH 8.0
儲存類別代碼
12 - Non Combustible Liquids
水污染物質分類(WGK)
nwg
分析證明 (COA)
輸入產品批次/批號來搜索 分析證明 (COA)。在產品’s標籤上找到批次和批號,寫有 ‘Lot’或‘Batch’.。
我們的科學家團隊在所有研究領域都有豐富的經驗,包括生命科學、材料科學、化學合成、色譜、分析等.
聯絡技術服務