H5000
Hexokinase from Saccharomyces cerevisiae
Type III, lyophilized powder, ≥25 units/mg protein (biuret)
Synonym(s):
ATP:D-Hexose-6-phosphotransferase, Hexokinase from yeast
About This Item
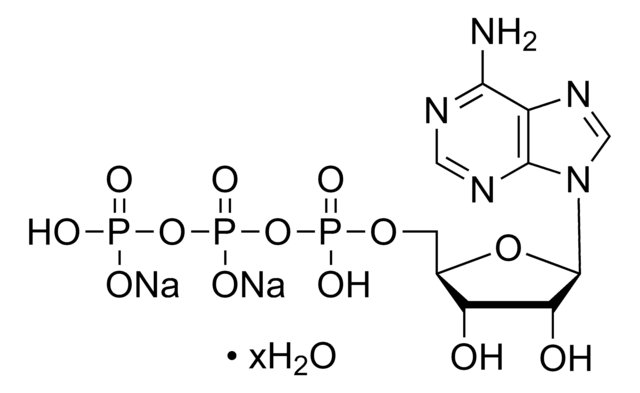
Recommended Products
type
Type III
form
lyophilized powder
specific activity
≥25 units/mg protein (biuret)
mol wt
dimer 110 kDa
foreign activity
glucose-6-phosphate dehydrogenase and phosphoglucose isomerase ≤10%
storage temp.
−20°C
Gene Information
bakers yeast ... HXK1(850614) , HXK2(852639)
Looking for similar products? Visit Product Comparison Guide
Biochem/physiol Actions
The rate of phosphorylation varies with different hexoses (pH 7.5, 30 °C).
D-fructose KM: 0.33 mM
D-glucose KM: 0.12 mM
D-mannose KM: 0.05 mM
Yeast hexokinase exists as two similar isoforms, PI and PII (A and B), with isoelectric points of 5.25 and 4, respectively.
Molecular Weight: ~ 54 kDa (monomer)
~110 kDa (dimer)
Optimal pH: 7.5 to 9.0
Extinction Coefficient: E1% = 8.85 (PI) and 9.47 (PII) at 280 nm
Activators: Hexokinase requires Mg2+ ions (KM = 2.6 mM) for activity. Hexokinase is activated by catecholamines and related compounds.
Inhibitors: sorbose-1-phosphate, polyphosphates, 6-deoxy-6-fluoroglucose, 2-C-hydroxy-methylglucose, xylose, lyxose, and thiol reactive compounds (Hg2+ and 4-chloromercuribenzoate)
Unit Definition
Physical form
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Personal Protective Equipment
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service