RNAINH-RO
Roche
Protector RNase Inhibitor
Synonym(s):
protector, rnase inhibitor
About This Item
Recommended Products
Assay
>95% (SDS-PAGE)
Quality Level
mol wt
~50 kDa
packaging
pkg of 10,000 U (03335402001 [5 x 2,000 U])
pkg of 2,000 U (03335399001)
manufacturer/tradename
Roche
parameter
25-55 °C optimum reaction temp.
pH range
5.0-9.0
storage temp.
−20°C
General description
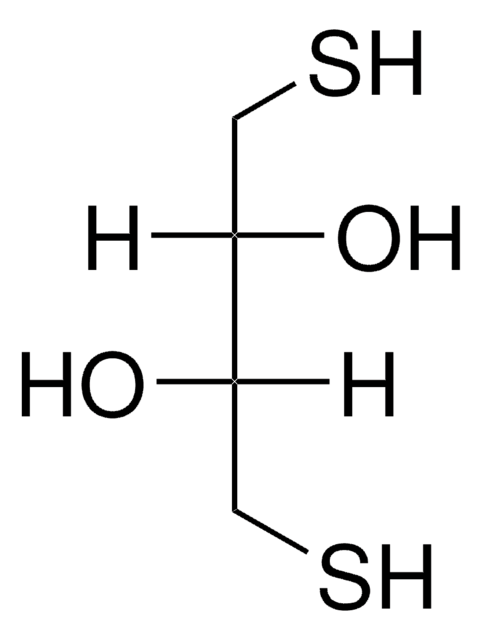
Protector RNase Inhibitor (40 U/μl) in storage buffer: 20 mM HEPES-KOH, 50 mM KCl, 8 mM dithiothreitol, 50% glycerol (v/v), pH approximately 7.6 (+4°C).
- RNase A
- RNase B
- RNase T2
- protect mRNA during cDNA synthesis reactions, RT-PCR (in conventional thermal cyclers and qPCR systems), or in vitro transcription/translation reactions
- protect viral RNA during in vitro virus replication
- inhibit RNases during RNA isolation and purification
- be used in RNase protection assays
- help prepare RNase-free antibodies
Specificity
RNase H is not inhibited by Protector RNase Inhibitor.
Heat inactivation: > 65 °C
Application
- Protect mRNA in cDNA synthesis reactions, RT-PCR (in conventional thermal cyclers and qPCR systems), e.g., with the LightCycler® Instruments, in vitro transcription/translation system, RNase Protection Assay, in vitro RNA synthesis
- Protect viral RNA during in vitro virus replication
- Inhibit RNases during RNA isolation and purification
- Help prepare RNase-free antibodies
- Synthesize RNA probes for in situ hybridization
Features and Benefits
- Rely on a thermostable RNase inhibitor: Protector RNase Inhibitor remains stable even when using thermostable reverse transcriptases such as Transcriptor Reverse Transcriptase.
- Benefit from a wide range of reaction conditions: Protector RNase Inhibitor is active at pH 5.0 to 9.0 and at temperatures between +25°C to +55°C (partial activity is still measureable at +60°C).
- Eliminate interference in different RNA analysis applications: Maintain performance even when adding at 16-fold higher than the standard concentration.
- Insist on a highly-purified preparation: Each batch is function tested using techniques such as quantitative RT-PCR to ensure the absence of endonucleases, ribonucleases, or nicking activity, according to the current quality control procedures.
Quality
Test buffer: 50 mM Tris-HCl, 10 mM MgCl2, 0.1 mM EDTA, 7 mM β-Mercaptoethanol; pH 7.5 (+37°C).
Absence of endonucleases: 1 μg EcoR I/Hind III*-fragments of λDNA is incubated with Protector RNase Inhibitor in 50 μl test buffer at +37°C for 1 hour. The amount of inhibitor that causes no alteration in the banding pattern is stated under "Endo".
Absence of nicking activity: 1 μg supercoiled pBR322 DNA is incubated with Protector RNase Inhibitor in 50 μl test buffer at +37°C for 1 hour. The amount of inhibitor that causes no relaxation of supercoiled DNA is stated under "Nick. Act.".
Absence of ribonuclease (1): 5 μg of MS2 RNA is incubated with Protector RNase Inhibitor for 1 hour at +37°C in a final volume of 50 μl. The amount of inhibitor that causes no degradation of MS2 RNA is stated under "RNase 1".
Absence of ribonuclease (2): 5 μg of MS2 RNA is incubated with Protector RNase Inhibitor for 1 hour at +37°C, then 10 minutes at +65°C in a final volume of 50 μl. The amount of inhibitor that causes no degradation of MS2 RNA is stated under "RNase 2".
Unit Definition
Unit Assay: Activity is measured according to Blackburn as ability to inhibit hydrolysis of cyclic cytidine-2′ : 3′-monophosphoric acid. Under assay conditions, 200 U of Protector RNase Inhibitor inhibits 50% of the activity of 1 μg RNase A.
Volume Activity: 40 U/μl
Preparation Note
- 5 to 10 U in One-step RT-PCR
- 25 to 50 U in Two step RT-PCR
- 20 U for in vitro transcription
of inhibitor did not interfere with RT-PCR.)
Other Notes
Source: Rat lung; Product is produced recombinant in E. coli
Isoelectric Point: pH 4.5
Temperature where Protector RNase Inhibitor is active: 25°C to +55°C; partial activity is still measurable at +60°C
Inactivation: Under severe denaturizing conditions (temperatures above +65°C), the inhibitor is inactivated
Bioburden: <50 cfu/ml
DNA Content: <100 pg/mg
Legal Information
This product is optimized for use in the polymerase chain reaction (PCR) covered by patents owned by F. Hoffmann-La Roche Ltd ("Roche"). No license under these patents to use the PCR Process is conveyed expressly or by implication to the purchaser by the purchase of this product. A license to use the PCR Process for certain research and development activities accompanies the purchase of certain Roche, Applied Biosystems or other licensed suppliers′ reagents when used in conjunction with an authorized thermal cycler, or is available from Applied Biosystems. Diagnostic purposes require a license from Roche. Further information on purchasing licenses may be obtained by contacting the Director of Licensing at Applied Biosystems, 850 Lincoln Centre Drive, Foster City, California 94404, USA.
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 1
Flash Point(F)
does not flash
Flash Point(C)
does not flash
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Protocols
The addition of Protector RNase Inhibitor to cell pellets is not recommended. Protector RNase Inhibitor as a 50kDa protein will most probably not permeate into intact cells. If the cell pellet is immediately frozen at -80 °C, this will be sufficient for stabilizing RNA. At -80 °C, Protector RNase Inhibitor is inactive.
Related Content
Polymerase chain reaction (PCR) is a technique for amplifying nucleic acid molecules and is commonly used in many applications, including RT-PCR, hot start PCR, end point PCR and more.
Polymerase chain reaction (PCR) is a technique for amplifying nucleic acid molecules and is commonly used in many applications, including RT-PCR, hot start PCR, end point PCR and more.
Polymerase chain reaction (PCR) is a technique for amplifying nucleic acid molecules and is commonly used in many applications, including RT-PCR, hot start PCR, end point PCR and more.
Polymerase chain reaction (PCR) is a technique for amplifying nucleic acid molecules and is commonly used in many applications, including RT-PCR, hot start PCR, end point PCR and more.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service