All Photos(2)
About This Item
Empirical Formula (Hill Notation):
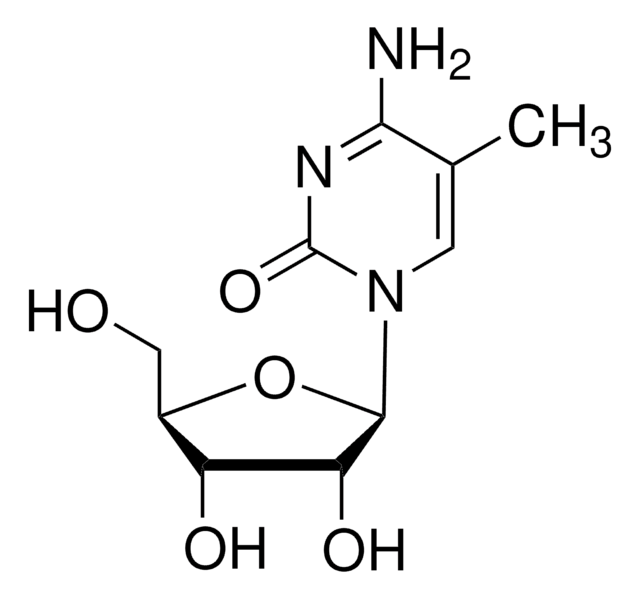
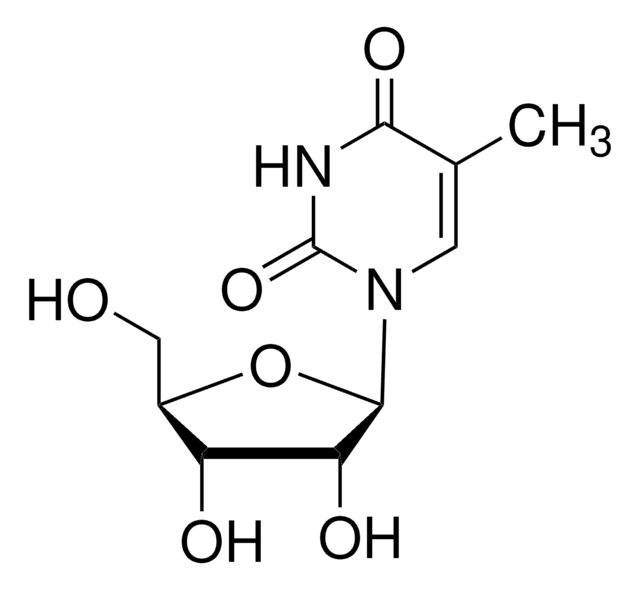
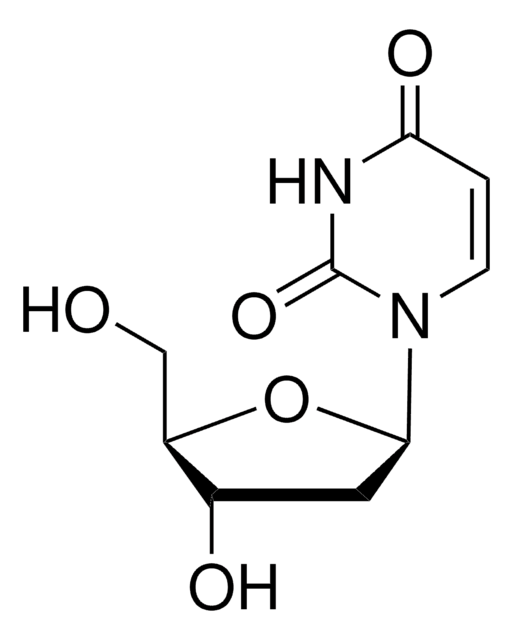
C10H14N2O6
CAS Number:
Molecular Weight:
258.23
EC Number:
MDL number:
UNSPSC Code:
41106305
PubChem Substance ID:
NACRES:
NA.22
Recommended Products
Assay
97%
mp
183-184 °C (lit.)
SMILES string
CC1=CN([C@@H]2O[C@H](CO)[C@@H](O)[C@H]2O)C(=O)NC1=O
InChI
1S/C10H14N2O6/c1-4-2-12(10(17)11-8(4)16)9-7(15)6(14)5(3-13)18-9/h2,5-7,9,13-15H,3H2,1H3,(H,11,16,17)/t5-,6-,7-,9-/m1/s1
InChI key
DWRXFEITVBNRMK-JXOAFFINSA-N
Looking for similar products? Visit Product Comparison Guide
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Personal Protective Equipment
dust mask type N95 (US), Eyeshields, Gloves
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Gregory E R Gordon et al.
Journal of biotechnology, 151(1), 108-113 (2010-11-30)
This paper describes a high yielding coupled enzymatic reaction using Bacillus halodurans purine nucleoside phosphorylase (PNP) and E. coli uridine phosphorylase (UP) for synthesis of 5-methyluridine (5-MU) by transglycosylation. Key parameters such as reaction temperature, pH, reactant loading, reactor configuration
S Wang et al.
Bioorganic & medicinal chemistry, 5(6), 1043-1050 (1997-06-01)
In DNA triple helices, methylation at C-5 of thymine or cytosine is reported to have similar stabilizing effects for both bases. Here we show, however, that methylation of the same positions in RNA triplexes has distinctly different effects than in
B Felden et al.
The EMBO journal, 17(11), 3188-3196 (1998-06-26)
Escherichia coli tmRNA functions uniquely as both tRNA and mRNA and possesses structural elements similar to canonical tRNAs. To test whether this mimicry extends to post-transcriptional modification, the technique of combined liquid chromatography/ electrospray ionization mass spectrometry (LC/ESIMS) and sequence
Sanjay Agarwalla et al.
The Journal of biological chemistry, 277(11), 8835-8840 (2002-01-10)
An Escherichia coli open reading frame, ygcA, was identified as a putative 23 S ribosomal RNA 5-methyluridine methyltransferase (Gustafsson, C., Reid, R., Greene, P. J., and Santi, D. V. (1996) Nucleic Acids Res. 24, 3756-3762). We have cloned, expressed, and
Ya-Ming Hou et al.
FEBS letters, 584(2), 278-286 (2009-12-01)
Methylation of tRNA on the four canonical bases adds structural complexity to the molecule, and improves decoding specificity and efficiency. While many tRNA methylases are known, detailed insight into the catalytic mechanism is only available in a few cases. Of
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service