推荐产品
一般說明
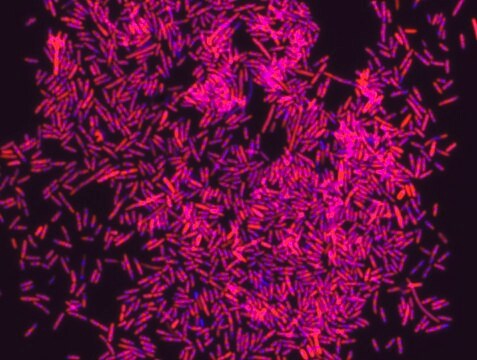
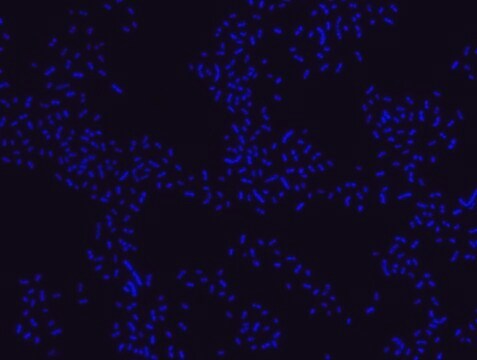
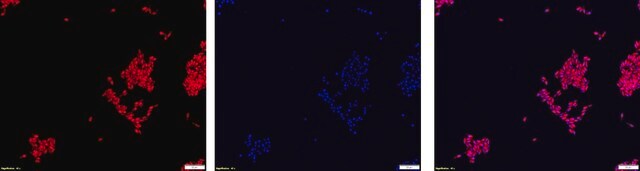
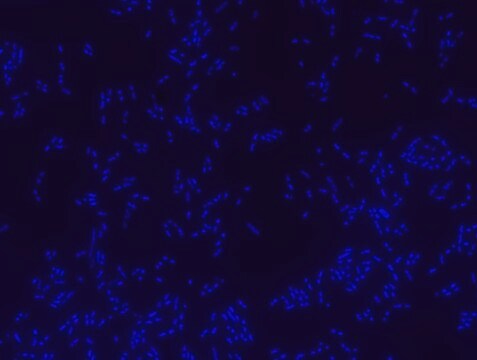
Fluorescent In Situ Hybridization technique (FISH) is based on the hybridization of fluorescent labeled oligonucleotide probe to a specific complementary DNA or RNA sequence in whole and intact cells. Microbial FISH allows the visualization, identification and isolation of bacteria due to recognition of ribosomal RNA also in unculturable samples.
FISH technique can serve as a powerful tool in the microbiome research field by allowing the observation of native microbial populations in diverse microbiome environments, such as samples from human origin (blood and tissue), microbial ecology (solid biofilms and aquatic systems) and plants.
Prokaryotic single cell life forms are divided into two domains, called Bacteria and Archaea, originally categorized as Eubacteria and Archaebacteria. However both terms, Eubacteria and Bacteria are still being used in microbiology. Eubacteria probe recognizes most bacteria as it is complementary to a portion of 16S rRNA found in almost all bacteria.,
FISH technique was successfully used to identify different bacteria with the universal bacterial probe in various samples such as, pure culture (as described in the figure legends), blood cultures,, periapical tooth lesions12, saliva13, biofilms from voice prostheses14, subgingival biofilm15, aortic wall tissue16, buccal epithelial cells, pure culture and cell culture17, intestine tissue embedded in paraffin18, necrotizing fasciitis and pure culture19, colon sections embedded in paraffin20,21, cancer tissues22,23, environmental samples24 and gut of the medicinal leech25. The probe can also be used for combined technique of FISH and Flow cytometric analysis. 9,26,27
It is strongly recommended to include positive and negative controls in FISH assays to ensure specific binding of the probe of interest and appropriate protocol conditions. We offer positive (MBD0032/33) and negative (MBD0034/35) control probes, that accompany the specific probe of interest.
FISH technique can serve as a powerful tool in the microbiome research field by allowing the observation of native microbial populations in diverse microbiome environments, such as samples from human origin (blood and tissue), microbial ecology (solid biofilms and aquatic systems) and plants.
Prokaryotic single cell life forms are divided into two domains, called Bacteria and Archaea, originally categorized as Eubacteria and Archaebacteria. However both terms, Eubacteria and Bacteria are still being used in microbiology. Eubacteria probe recognizes most bacteria as it is complementary to a portion of 16S rRNA found in almost all bacteria.,
FISH technique was successfully used to identify different bacteria with the universal bacterial probe in various samples such as, pure culture (as described in the figure legends), blood cultures,, periapical tooth lesions12, saliva13, biofilms from voice prostheses14, subgingival biofilm15, aortic wall tissue16, buccal epithelial cells, pure culture and cell culture17, intestine tissue embedded in paraffin18, necrotizing fasciitis and pure culture19, colon sections embedded in paraffin20,21, cancer tissues22,23, environmental samples24 and gut of the medicinal leech25. The probe can also be used for combined technique of FISH and Flow cytometric analysis. 9,26,27
It is strongly recommended to include positive and negative controls in FISH assays to ensure specific binding of the probe of interest and appropriate protocol conditions. We offer positive (MBD0032/33) and negative (MBD0034/35) control probes, that accompany the specific probe of interest.
應用
Eubacteria FISH probe - ATTO488 is suitable to use as a probe for fluorescence in situ hybridization (FISH) to recognize Eubacteria cells .
特點和優勢
- Visualize, identify and isolate bacteria cells.
- Observe native bacteria cell populations in diverse microbiome environments.
- Specific, sensitive and robust identification of bacteria cells in mixed microorganism population.
- Specific, sensitive and robust identification even when bacteria are in low abundance in the sample.
- FISH can complete PCR based detection methods by avoiding contaminant bacteria detection.
- Provides information on bacteria morphology and allows to study biofilm architecture.
- Identify various bacteria in environmental and clinical samples such as, formalin-fixed paraffin-embedded (FFPE) samples, blood cultures, saliva and more.
- The ability to detect bacteria in its natural habitat is an essential tool for studying host-microbiome interaction.
儲存類別代碼
12 - Non Combustible Liquids
水污染物質分類(WGK)
nwg
閃點(°F)
Not applicable
閃點(°C)
Not applicable
Lionel Rigottier-Gois et al.
Systematic and applied microbiology, 26(1), 110-118 (2003-05-16)
Bacteroides is a predominant group of the faecal microbiota in healthy adults. To investigate the species composition of Bacteroides by fluorescent in situ hybridisation (FISH) combined with flow cytometry, we developed five species-specific probes targeting the 16S rRNA. Probes were
Yu-Yuan Li et al.
World journal of gastroenterology, 22(11), 3227-3233 (2016-03-24)
To investigate Fusobacterium nucleatum (F. nucleatum) abundance in colorectal cancer (CRC) tissues and its association with CRC invasiveness in Chinese patients. The resected cancer and adjacent normal tissues (10 cm beyond cancer margins) from 101 consecutive patients with CRC were
Pia T Sunde et al.
Microbiology (Reading, England), 149(Pt 5), 1095-1102 (2003-05-02)
Whether micro-organisms can live in periapical endodontic lesions of asymptomatic teeth is under debate. The aim of the present study was to visualize and identify micro-organisms within periapical lesions directly, using fluorescence in situ hybridization (FISH) in combination with epifluorescence
Dorothee Maria Gescher et al.
International journal of antimicrobial agents, 32 Suppl 1, S51-S59 (2008-08-23)
Sepsis is a life-threatening disease with a high mortality rate. Rapid identification of blood culture isolates plays a crucial role in adequate antimicrobial therapy in sepsis patients. To accelerate microbiological diagnosis, a comprehensive panel of oligonucleotide probes for fluorescence in
Michele A Maltz et al.
Frontiers in microbiology, 5, 151-151 (2014-05-27)
There are trillions of microbes found throughout the human body and they exceed the number of eukaryotic cells by 10-fold. Metagenomic studies have revealed that the majority of these microbes are found within the gut, playing an important role in
我们的科学家团队拥有各种研究领域经验,包括生命科学、材料科学、化学合成、色谱、分析及许多其他领域.
联系技术服务部门