推荐产品
一般說明
Metagenomics analysis looks at all DNA that has been isolated directly from given single samples (e.g. environmental samples, biological organisms). Metagenomics allows for the investigation of microbes that exist in any environment (including extreme environments), and which have been historically difficult to isolate, culture, and study. Metagenomics has revealed the existence of novel microbial species. Applications of metagenomic studies include public health data analysis, discovery of novel proteins, enzymes and natural products, environmental studies, and agricultural investigations.
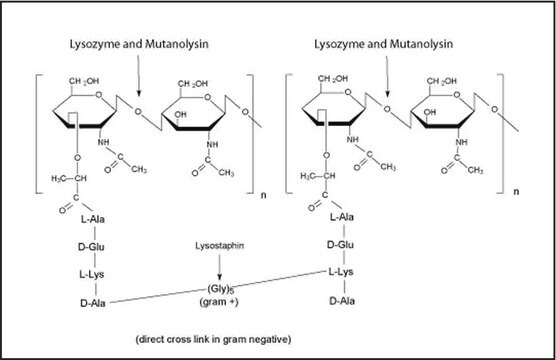
MetaPolyzyme products (MAC4L and MAC4LDF) are based on a multi-lytic enzyme mixture formulated for effective lysis of Microbiome samples from extreme environments. Originally developed by Scott Tighe for use in microbiome and DNA extraction efficiency studies, the products were evaluated and developed in consultation and collaboration with the Association of Biomolecular Resource Facilities (ABRF) Metagenomics and Microbiome Research Group (MMRG; formerly the Metagenomics Research Group, MGRG).1-4 Several publications have cited use of MetaPolyzyme.5-7
MetaPolyzyme products (MAC4L and MAC4LDF) are based on a multi-lytic enzyme mixture formulated for effective lysis of Microbiome samples from extreme environments. Originally developed by Scott Tighe for use in microbiome and DNA extraction efficiency studies, the products were evaluated and developed in consultation and collaboration with the Association of Biomolecular Resource Facilities (ABRF) Metagenomics and Microbiome Research Group (MMRG; formerly the Metagenomics Research Group, MGRG).1-4 Several publications have cited use of MetaPolyzyme.5-7
應用
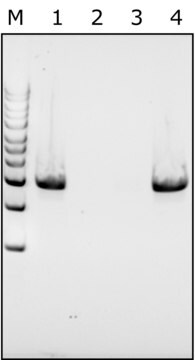
Our experts have shown that using MetaPolyzyme, DNA free in a microbial study can increase final DNA concentration by approximately 2X, and increase the number of taxa identified. It is useful for low biomass samples for lysing cells without contributing to DNA background contamination. Learn more about the experiments and data by reviewing our technical article – MetaPolyzyme, DNA Free Cell Lysis Enzymes for Microbiome Workflows.
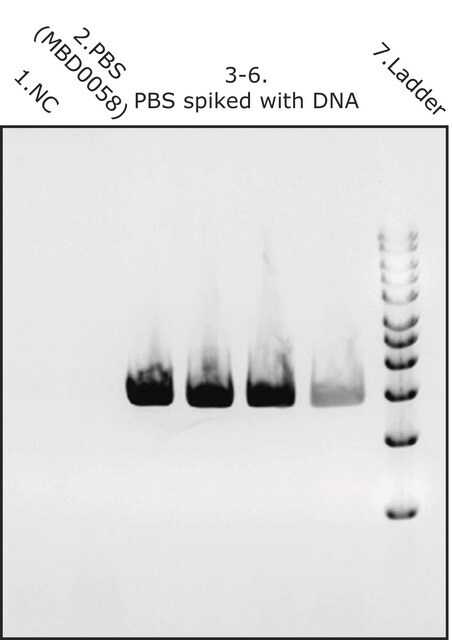
The study of microbial communities has been revolutionized in recent years by the widespread adoption of culture independent analytical techniques such as 16S rRNA gene sequencing and metagenomics. Since DNA contamination during sample preparation is a major problem of these sequence-based approaches, DNA extraction reagents free of DNA contaminants are essential. MetaPolyzyme, DNA free, undergoes strict quality control testing to ensure the absence of detectable levels of contaminating microbial DNA using 35 cycles PCR amplification of 16S and 18S rDNA using universal primer sets.
The need for DNA free reagents led to the development of MetaPolyzyme, DNA free, which is essential when there is a need to minimize microbial DNA contamination from reagents during microbiome studies.
The study of microbial communities has been revolutionized in recent years by the widespread adoption of culture independent analytical techniques such as 16S rRNA gene sequencing and metagenomics. Since DNA contamination during sample preparation is a major problem of these sequence-based approaches, DNA extraction reagents free of DNA contaminants are essential. MetaPolyzyme, DNA free, undergoes strict quality control testing to ensure the absence of detectable levels of contaminating microbial DNA using 35 cycles PCR amplification of 16S and 18S rDNA using universal primer sets.
The need for DNA free reagents led to the development of MetaPolyzyme, DNA free, which is essential when there is a need to minimize microbial DNA contamination from reagents during microbiome studies.
特點和優勢
Multi-lytic enzyme mixture designed for effective lysis of microbiome samples
DNA-free formulation minimizes contamination
Ideal for low biomass samples
Strict quality control testing ensures absence of detectable levels of contaminating microbial DNA
Increases final DNA concentration and number of taxa identified
DNA-free formulation minimizes contamination
Ideal for low biomass samples
Strict quality control testing ensures absence of detectable levels of contaminating microbial DNA
Increases final DNA concentration and number of taxa identified
成分
The enzymes in MetaPolzyme, DNA free are:
All the enzymes are individually tested for absence of contaminating microbial DNA using 16S and 18S PCR amplification
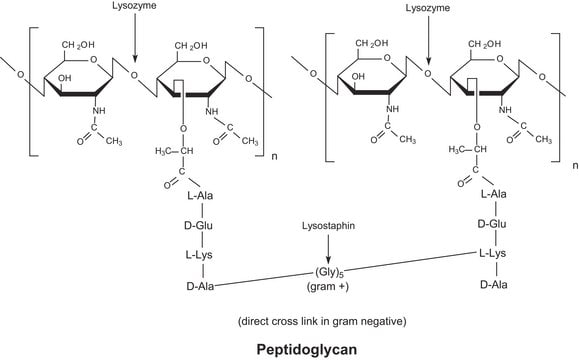
- Mutanolysin
- Achromopeptidase
- Chitinase
- Lyticase
- Lysostaphin
- Lysozyme
All the enzymes are individually tested for absence of contaminating microbial DNA using 16S and 18S PCR amplification
訊號詞
Danger
危險聲明
危險分類
Resp. Sens. 1
儲存類別代碼
11 - Combustible Solids
水污染物質分類(WGK)
WGK 3
閃點(°F)
Not applicable
閃點(°C)
Not applicable
我们的科学家团队拥有各种研究领域经验,包括生命科学、材料科学、化学合成、色谱、分析及许多其他领域.
联系技术服务部门