I7634
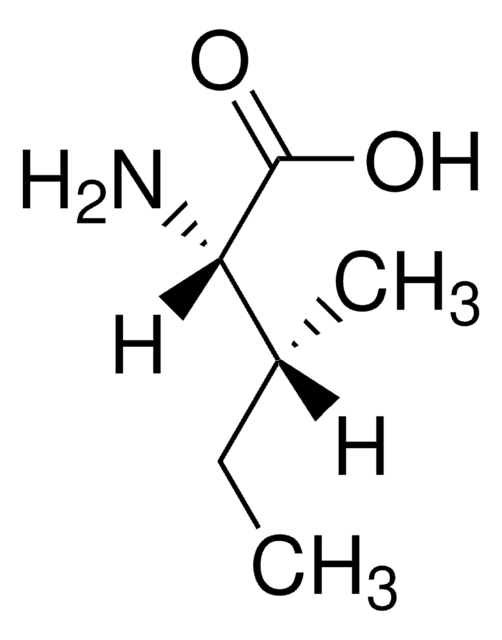
D-Isoleucine
≥98% (TLC)
Synonym(s):
(2R, 3R)-2-Amino-3-methylpentanoic acid
About This Item
Recommended Products
product name
D-Isoleucine, ≥98% (TLC)
Quality Level
Assay
≥98% (TLC)
form
powder
impurities
≤10% allo-isomer
color
white
application(s)
cell analysis
SMILES string
CC[C@@H](C)[C@@H](N)C(O)=O
InChI
1S/C6H13NO2/c1-3-4(2)5(7)6(8)9/h4-5H,3,7H2,1-2H3,(H,8,9)/t4-,5-/m1/s1
InChI key
AGPKZVBTJJNPAG-RFZPGFLSSA-N
Looking for similar products? Visit Product Comparison Guide
Related Categories
Application
- Metabolic Profiling of Urinary Chiral Amino-Containing Biomarkers for Gastric Cancer Using a Sensitive Chiral Chlorine-Labeled Probe by HPLC-MS/MS.: This study by Huang et al. (2021) explores the use of D-Isoleucine as a biomarker for gastric cancer, utilizing advanced metabolic profiling techniques with HPLC-MS/MS to detect chiral amino-containing compounds in urine samples (Huang et al., 2021).
- Isopenicillin N Synthase Binds δ-(L-α-aminoadipoyl)-L-cysteinyl-D-thia-allo-isoleucine Through Both Sulfur Atoms.: Clifton et al. (2011) describe the binding mechanism of Isopenicillin N synthase with a compound involving D-Isoleucine, highlighting its role in antibiotic biosynthesis and enzyme interaction studies (Clifton et al., 2011).
- The Total Structure of the Antibiotic Longicatenamycin.: Shiba and Mukunoki (1975) determined the complete structure of the antibiotic longicatenamycin, which includes D-Isoleucine as a crucial component, contributing to understanding its antibacterial properties (Shiba and Mukunoki, 1975).
Biochem/physiol Actions
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Chromatograms
application for HPLCOur team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service