SeqPlex™-I Whole Transcriptome Amplification Kits for Next-Generation Sequencing
- What is Whole Transcriptome Amplification?
- Whole Transcriptome Library Preparation using SeqPlex™-I WTA Kits
- SeqPlex™-I Whole Transcriptome Amplification and Adapter-Ligation Workflow Comparison
- Total Gene Identification for WTA Workflows with and without rRNA Depletion
- Whole Transcriptome Amplification Non-Coding Gene Expression Analysis
- Whole Transcriptome Amplification Kits and Reagent Ordering
What is Whole Transcriptome Amplification?
Whole transcriptome amplification (WTA) is a method often used to amplify small quantities of reverse transcribed RNA, or degraded RNA for direct use in Next-Generation Sequencing (NGS) workflows. To facilitate your WTA workflow needs, we offer a collection of WTA products for amplification of total RNA from a variety of organisms. All WTA kits are designed specifically for low-input templates and low-quality nucleic acids, such as samples isolated from FFPE tissues or RNA immunoprecipitation (RIP). Each of our WTA kits are optimized for specific applications. The Transplex® WTA kit is ideal for microarrays, while the SeqPlex™ WTA kit is designed for use with any NGS instrument systems after primer removal and ligation of instrument-specific adaptors. Finally, the SeqPlex™-I WTA library preparation kit has a finishing step to add Illumina® adapters, producing libraries that are ready for direct input onto Illumina® NGS flow cells.
Whole Transcriptome Library Preparation using SeqPlex™-I WTA Kits
All our WTA kits follow the same general principle for random library generation, but the workflows diverge after the library amplification to be compatible with the desired analysis method (Figure 1). First, primers, which are composed of a poly(T) or a semi-degenerate 3’ end and a universal 5’ end, land randomly on input RNA. After annealing, the primers are extended, displacing adjacent amplification products. The displaced amplification products serve as templates for subsequent second-strand synthesis, resulting in a library of double-stranded products containing the universal 5’ sequence and its reverse complement at both ends.
Next, the double-stranded products are amplified by single primer PCR via the proprietary universal 5’ sequence. TransPlex® kit workflows are complete after the universal PCR step. Use of SeqPlex™ kits are followed by a primer removal step to eliminate the universal primers, resulting in products that are ready to enter any adapter ligation workflow for NGS. Finally, SeqPlex™-I kit workflows have an additional amplification step to replace the universal primer sequence with Illumina® adapters, producing libraries that are ready for direct input onto Illumina® NGS flow cells.
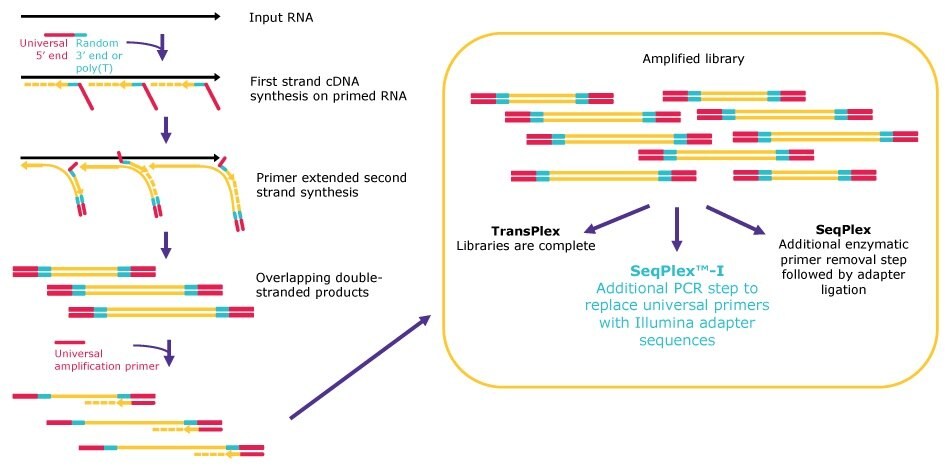
Figure 1.Whole transcriptome amplification workflows.
The SeqPlex™-I WTA kit (SEQRI) provides several workflow efficiencies compared to other library preparation methods. Unlike traditional adapter ligation methods, the SeqPlex™-I workflow does not require fragmentation, end repair, or intermediate bead cleanup steps, which saves valuable hands-on time. Additionally, ribosomal RNA (rRNA) depletion is not required to achieve maximal transcriptome coverage using SeqPlex™-I kits.
SeqPlex™-I Whole Transcriptome Amplification and Adapter-Ligation Workflow Comparison
The SeqPlex™-I WTA workflow was compared to an adapter-ligation based workflow, with both workflows tested with and without a prior ribosomal RNA depletion step. The resulting libraries were analyzed for ribosomal RNA content, coding gene content, and non-coding RNA content. Figure 2 shows a comparison of ribosomal RNA reads detected in the libraries prepared with the SeqPlex™-I kit or ligation-based methods with or without a ribosomal RNA (rRNA) depletion process.
For the ligation-based library preps, without an rRNA depletion process, 90% of reads were aligned to rRNA while only 10% of reads could be used for the analysis of other RNAs. When ligation was paired with an rRNA depletion process, rRNA was efficiently removed and most of the reads could be used for transcriptome analyses, though at least 10 ng of input RNA was required for the library prep. With a 1 ng input, a library was not successfully produced. Since the proprietary semi-degenerate primers preferably amplify the non-ribosomal RNA, only about 38% of reads were aligned to rRNA with libraries produced with the SeqPlex™-I kit, leaving 62% of reads to be usable for transcriptome analysis. When paired with rRNA depletion, SeqPlex™-I yielded 3% of reads aligned to rRNA, with almost 97% of the reads available for transcriptome analysis.
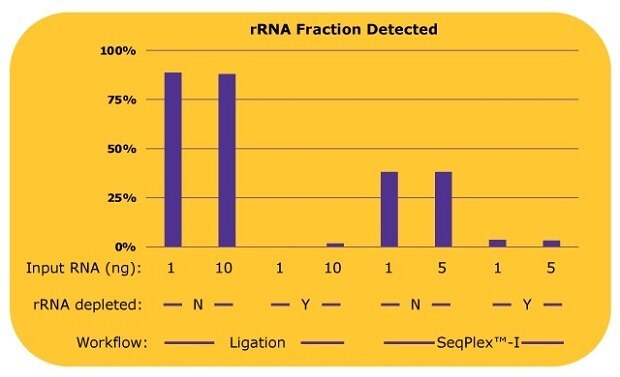
Figure 2.Percentages of rRNA reads in each library with differing library preparation workflows. Whole transcriptome libraries were prepared by a ligation-based method or the SeqPlex™-I WTA kit from 1 ng or 10 ng of universal human reference total RNA. Sequencing was performed on an Illumina® NextSeq 550 System with 50 million reads per library. The sequencing data were aligned to RefSeq (hg38) with BWA and the percentages of the rRNA reads out of total reads were calculated. The library was not able to be prepared with 1 ng of RNA by the ligation-based method after the rRNA depletion process.
Total Gene Identification for WTA Workflows with and without rRNA Depletion
Additional analysis shows the number of genes identified by each library (Figure 3). With the ligation-based library prep, about 10,000 genes were identified without an rRNA depletion process, since only 10% of the total reads, 5 million out of 50 million reads were derived from non-rRNA. With an rRNA depletion process and 10 ng input of RNA, the number of the identified genes from a ligation-based workflow increased to over 18,000. With the SeqPlex™-I library preparation, 14,000 genes were identified with 1 ng of input RNA, and 23,000 genes were identified with 5 ng of input RNA without the rRNA depletion process. Similar amounts or more genes were identified with 5 ng input of RNA without an rRNA depletion process with the SeqPlex-I library preparation. An additional rRNA depletion process for the SeqPlex™-I library preparation improved the identification of genes with 1 ng of input RNA but had no effect when using 5 ng of input RNA.
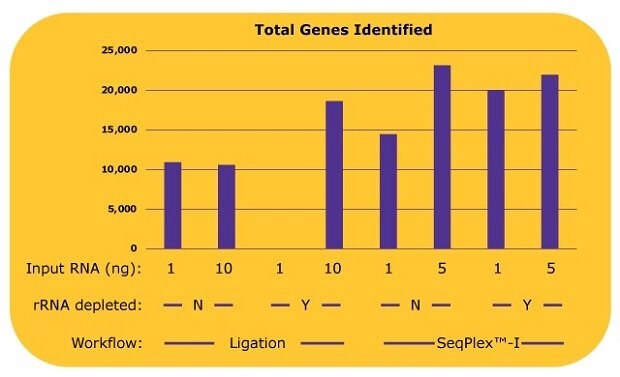
Figure 3.The number of genes identified in each library with differing library preparation workflows. Gene expression was analyzed with STAR aligner using the sequence data from the experiment as described in Figure 2. To count as “identified”, genes required a read count of >10.
Whole Transcriptome Amplification Non-Coding Gene Expression Analysis
The difference between the library preparation processes was even more pronounced when identifying non-coding RNA genes, whose copy numbers are low. With the ligation-based library prep, only 500 to 600 non-coding genes were identified without an rRNA depletion process. When an rRNA depletion was processed with 10 ng input of RNA, about 4,000 non-coding genes were identified. Alternatively, using the SeqPlex™-I process, approximately 2,400 non-coding genes were identified with 1 ng of input RNA, and about 7,400 non-coding genes were identified with 5 ng of input RNA without rRNA depletion processes (Figure 4). An additional rRNA depletion process for the SeqPlex™-I library preparation improved the identification of non-coding RNA genes to about 5,500 with 1 ng of input RNA; however, no improvements were observed using 5 ng of input RNA.
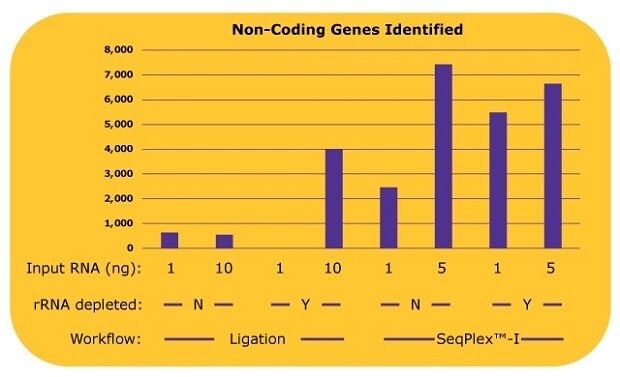
Figure 4.The number of non-coding genes identified in each library with differing library preparation workflows. The non-coding gene expressions were analyzed from the alignment data in Figure 3.
Whole Transcriptome Amplification Kits and Reagent Ordering
In summary, whole transcriptome amplification based NGS library preparation is a useful method to analyze gene expression. The SeqPlex™-I whole transcriptome amplification kit is designed specifically for low-input templates and low-quality nucleic acids and is compatible with Illumina® NGS instrumentation. Compared to adapter ligation, libraries created with SeqPlex™-I require fewer hands-on steps and can be more cost-effective when bypassing the need for ribosomal RNA depletion. In addition, more coding and non-coding genes can be identified with SeqPlex™-I kits than ligation-based library preparation methods from less than 5 ng of input sample. Explore all our whole transcriptome amplification kits and related reagents in the product selection below.
To continue reading please sign in or create an account.
Don't Have An Account?