Protein Pull-Down Techniques
The pull-down assay is designed to determine the interaction of two or more proteins. With affinity pull-down assays, a ″bait″ protein is tagged and captured on an immobilized ligand (support beads) by covalent attachment or through an affinity tag such as immobilized metal affinity chromatography (IMAC). Common bait protein fusion tags include Glutathione S-transferase (GST) and histidine-tagged proteins. The bait protein forms a complex which is then incubated with a protein source such as cell or tissue lysate. The proteins of interest are eluted from the beaded support through a series of washes and collected by centrifugation. The affinity pull-down method of purification and detection is different from immunoprecipitation (IP) and Co-IP assays in that it is not an antibody to antigen interaction. With all pull-down assays, the purified sample is often analyzed by SDS-PAGE, mass spectral analysis, and western blot detection for further downstream analysis.
Related Technical Articles
- Differential centrifugation and density gradient centrifugation are two types of centrifugal techniques for separating particles.CsCl gradient centrifugation, or Caesium chloride centrifugation is used to make solutions for the separation of RNA from DNA by density gradient centrifugation.
- Centrifugation enables the separation of particles by sedimentation. Learn how to separate particles using a centrifuge and how to use Stokes' law to calculate the velocity of sedimentation.
- Sera-Mag and Sera-Mag SpeedBeads provide cost effective magnetic bead separation technology for molecular biology applications, nucleic acid isolation, and research immunoassays.
- Agarose beads Vs. Magnetic beads in Chromatin Immunoprecipitation (ChIP)
- This page describes immunoprecipitation (immunoaffinity or pull-down techniques).
- See All (10)
Related Protocols
- To determine the molecular weights of protein antigens, to study protein/protein interactions, to determine specific enzymatic activity, to monitor protein post-translational modifications and to determine the presence and quantity of proteins.
- Immunoprecipitation Kit (Protein G) Protocol & Troubleshooting
- Immunoprecipitation Kit (Protein A) Protocol & Troubleshooting
- Sera-Mag™ SpeedBeads Magnetic Protein A/G-Particles Protocol
- Nanodisc technology offers a means to overcome some of the challenges associated with solubilizing membrane proteins. Nanodisc technology is applicable to a wide variety of membrane proteins and protein classes.
- See All (13)
Find More Articles and Protocols
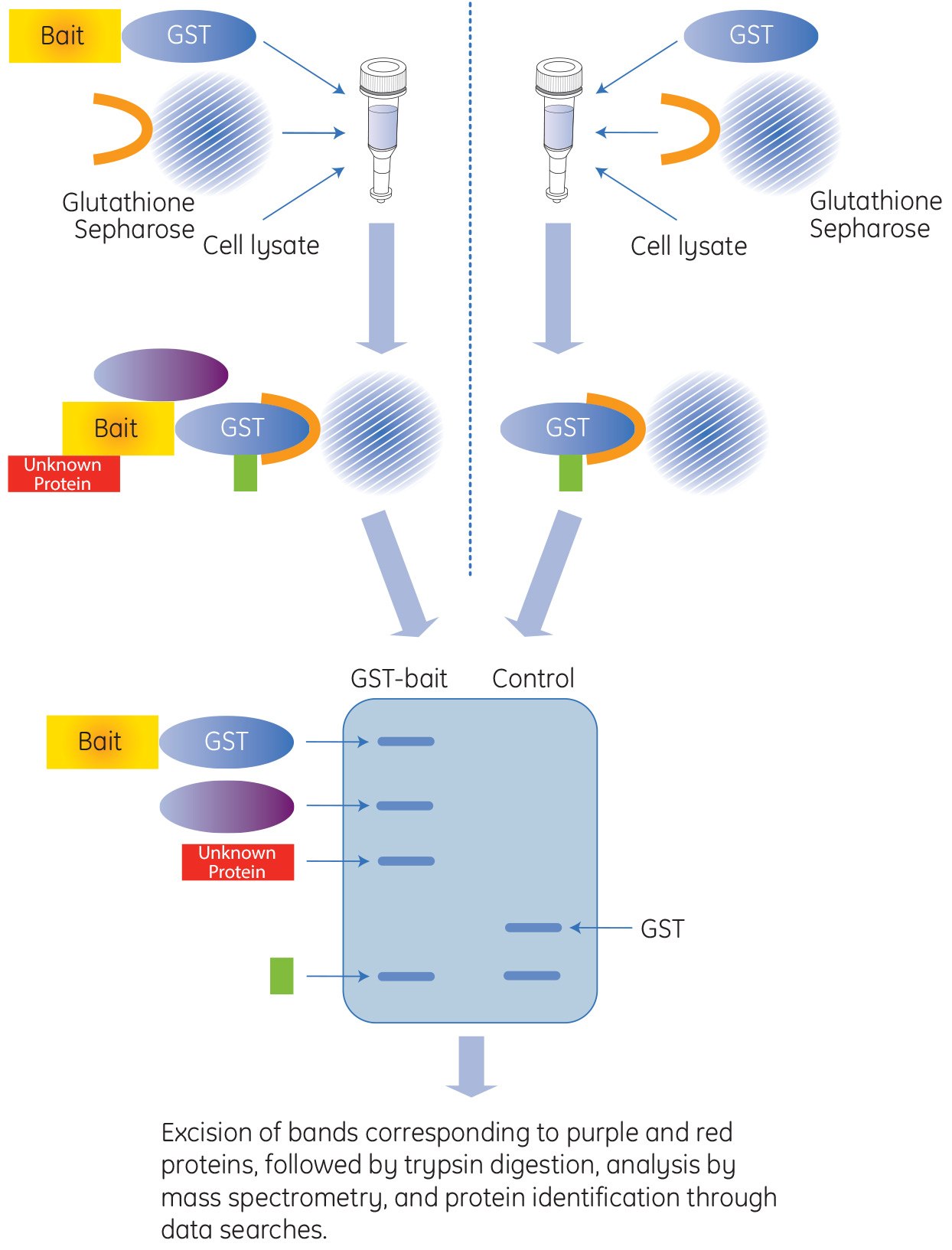
Affinity GST Pull-Down Applications
Glutathione S-transferase (GST) is a 211 amino acid protein that is found in most organisms. GST is often integrated into expression vectors to produce the fusion tag within E. coli protein expression systems. The GST-tag is a large protein tag, approximately 26 kDa, and can be expressed in bacteria, yeast, mammalian, and insect cells. The GST-tag is advantageous when protection of the recombinant protein from intracellular protease cleavage is required, and when there is a need to enhance the solubility of the tagged protein. In addition, the GST protein provides a high-affinity tag for easy purification and for pull-down experiments utilizing inexpensive affinity resins that are performed in mild elution conditions.
Co-Immunoprecipitation
The pull-down assay is ideal for verification of co-immunoprecipitation results and an excellent method of identifying unknown protein-protein interactions along with the activation status of specific proteins. Unlike affinity pull-down methods, IP uses antibodies to bind to and isolate antigens from complex biological samples.
Co-IP refers to the use of an antibody to bind an antigen within a multiprotein complex. The complex is then captured with the addition of protein A or protein G medium. The high affinity of protein A/G to the Fc region of polyclonal and monoclonal IgG-type antibodies makes them a critical component in the purification of the protein or protein complex of interest. The use of protein pull-down methods is an essential tool for investigating protein-protein interaction networks; however, the choice of the specific method will be determined by the researcher’s specific application need.
To continue reading please sign in or create an account.
Don't Have An Account?