C0483
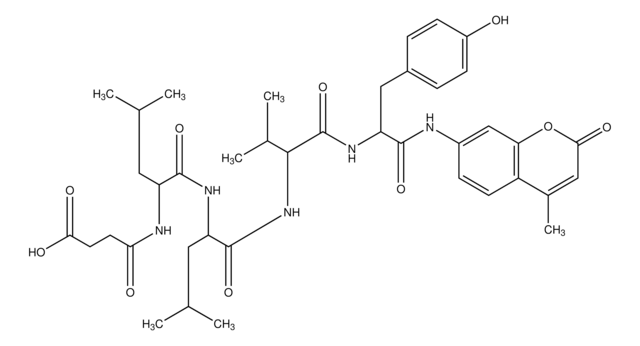
Z-Leu-Leu-Glu-7-amido-4-methylcoumarin
≥95%, solid
Synonym(s):
Cbz-Leu-Leu-Glu-AMC, Z-LLE-AMC
About This Item
Recommended Products
Quality Level
Assay
≥95%
form
solid
solubility
DMSO: soluble
storage temp.
−20°C
SMILES string
CC(C)C[C@H](NC(=O)OCc1ccccc1)C(=O)N[C@@H](CC(C)C)C(=O)N[C@@H](CCC(O)=O)C(=O)Nc2ccc3C(C)=CC(=O)Oc3c2
InChI
1S/C35H44N4O9/c1-20(2)15-27(38-34(45)28(16-21(3)4)39-35(46)47-19-23-9-7-6-8-10-23)33(44)37-26(13-14-30(40)41)32(43)36-24-11-12-25-22(5)17-31(42)48-29(25)18-24/h6-12,17-18,20-21,26-28H,13-16,19H2,1-5H3,(H,36,43)(H,37,44)(H,38,45)(H,39,46)(H,40,41)/t26-,27-,28-/m0/s1
InChI key
FOYHOBVZPWIGJM-KCHLEUMXSA-N
Amino Acid Sequence
General description
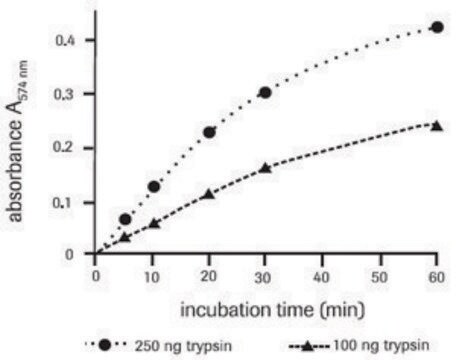
Application
Biochem/physiol Actions
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Personal Protective Equipment
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service