324725
Endoglycosidase F1, Elizabethkingia meningosepticum, Recombinant, E. coli
Endoglycosidase F1, Elizabethkingia meningosepticum, Recombinant, E. coli cleaves asparagine-linked or free oligomannose and hybrid. Suitable for deglycosylation of native proteins.
Sinónimos:
Endo-β-N-acetylglucosaminidase F1, Endo F1
Iniciar sesiónpara Ver la Fijación de precios por contrato y de la organización
About This Item
Productos recomendados
recombinant
expressed in E. coli
Quality Level
conjugate
(N-linked)
form
liquid
specific activity
≥16 units/mg protein
≥17 units/mL
manufacturer/tradename
Calbiochem®
storage condition
do not freeze
foreign activity
Proteases, none detected
shipped in
wet ice
storage temp.
2-8°C
Categorías relacionadas
General description
Note: 1 mU = 1 milliunit.
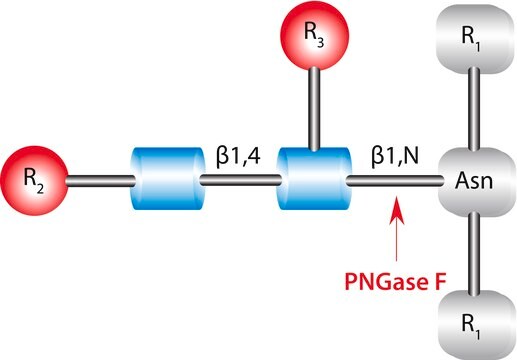
Recombinant, Elizabethkingia meningosepticum endoglycosidase F1 expressed in E. coli. Cleaves asparagine-linked or free oligomannose and hybrid, but not complex oligosaccharides. Core fucosylation reduces activity by 50 fold. Endo F1 will hydrolyze sulfate containing high mannose chains. It cleaves between the two N-acetylglucosamine residues in the diacetylchitobiose core of the oligosaccharide generating a truncated sugar molecule with one N-acetylglucosamine residue remaining on the asparagine. Less sensitive to protein conformation than N-Glycosidase F (Cat. No. 362185) and therefore is more suitable for deglycosylation of native proteins.
Warning
Toxicity: Standard Handling (A)
Unit Definition
One unit is defined as the amount of enzyme that will release N-linked oligosaccharides from 1.0 µmol denatured ribonuclease B per min at 37°C, pH 5.5.
Physical form
In 20 mM Tris-HCl, pH 7.5.
Other Notes
Tarentino, A.L., and Plummer, T.H. 1994. Methods Enzymol. 230, 44.
Tarentino, A.L., et al. 1992. J. Biol. Chem. 267, 3868.
Trimble, R.B., and Tarentino, A.L. 1991. J. Biol. Chem. 266, 1646.
Tarentino, A.L., et al. 1992. J. Biol. Chem. 267, 3868.
Trimble, R.B., and Tarentino, A.L. 1991. J. Biol. Chem. 266, 1646.
Legal Information
CALBIOCHEM is a registered trademark of Merck KGaA, Darmstadt, Germany
Storage Class
10 - Combustible liquids
wgk_germany
WGK 1
flash_point_f
Not applicable
flash_point_c
Not applicable
Certificados de análisis (COA)
Busque Certificados de análisis (COA) introduciendo el número de lote del producto. Los números de lote se encuentran en la etiqueta del producto después de las palabras «Lot» o «Batch»
¿Ya tiene este producto?
Encuentre la documentación para los productos que ha comprado recientemente en la Biblioteca de documentos.
A L Tarentino et al.
The Journal of biological chemistry, 267(6), 3868-3872 (1992-03-06)
A full-length insert for the endo-beta-N-acetylglucosaminidase (Endo) F1 gene was located on a 2,200-base pair EcoRI fragment of genomic DNA and cloned into the plasmid vector Bluescript. Transformed Escherichia coli cells expressed Endo F1 activity very well, but the enzyme
R B Trimble et al.
The Journal of biological chemistry, 266(3), 1646-1651 (1991-01-25)
Flavobacterium meningosepticum endo-beta-acetyl-glucosaminidase F preparations have been resolved by hydrophobic interaction chromatography on TSK-butyl resin into at least three activities designated endo F1, endo F2 and endo F3 each with a unique substrate specificity. The 32-kDa endo F1 protein is
Enzymatic deglycosylation of asparagine-linked glycans: purification, properties, and specificity of oligosaccharide-cleaving enzymes from Flavobacterium meningosepticum.
A L Tarentino et al.
Methods in enzymology, 230, 44-57 (1994-01-01)
Thapakorn Jaroentomeechai et al.
Nature communications, 13(1), 6325-6325 (2022-10-25)
The ability to reconstitute natural glycosylation pathways or prototype entirely new ones from scratch is hampered by the limited availability of functional glycoenzymes, many of which are membrane proteins that fail to express in heterologous hosts. Here, we describe a
Nuestro equipo de científicos tiene experiencia en todas las áreas de investigación: Ciencias de la vida, Ciencia de los materiales, Síntesis química, Cromatografía, Analítica y muchas otras.
Póngase en contacto con el Servicio técnico