790528P
Avanti
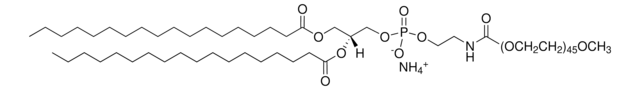
18:1 DGS-NTA
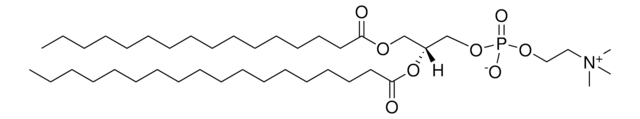
1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (ammonium salt), powder
Sinónimos:
1,2-di-(9Z-octadecenoyl)-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (ammonium salt); DOGS NTA
About This Item
Productos recomendados
assay
>99% (TLC)
form
powder
packaging
pkg of 1 × 5 mg (790528P-5mg)
manufacturer/tradename
Avanti Research™ - A Croda Brand 790528P
shipped in
dry ice
storage temp.
−20°C
Categorías relacionadas
General description
Application
- for generation of amphiphile monolayer
- in NTA-liposome preparation for procoagulant binding studies and to study its effect on factor FXII autoactivation
- as a liposome component for lipid bilayer preparation for surface plasmon resonance studies
Biochem/physiol Actions
Packaging
Legal Information
Storage Class
11 - Combustible Solids
Elija entre una de las versiones más recientes:
Certificados de análisis (COA)
Lo sentimos, en este momento no disponemos de COAs para este producto en línea.
Si necesita más asistencia, póngase en contacto con Atención al cliente
¿Ya tiene este producto?
Encuentre la documentación para los productos que ha comprado recientemente en la Biblioteca de documentos.
Nuestro equipo de científicos tiene experiencia en todas las áreas de investigación: Ciencias de la vida, Ciencia de los materiales, Síntesis química, Cromatografía, Analítica y muchas otras.
Póngase en contacto con el Servicio técnico
![18:1 DGS-NTA(Co) 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (Cobalt salt), powder](/deepweb/assets/sigmaaldrich/product/structures/203/482/b877a7a9-e8a9-4cec-99cf-b256a26d690c/640/b877a7a9-e8a9-4cec-99cf-b256a26d690c.png)