E1014
Benzonase® Nuclease
≥250 units/μL, ≥90% (SDS-PAGE), recombinant, expressed in E. coli, buffered aqueous glycerol solution
Synonym(s):
Endonuclease from Serratia marcescens
About This Item
Recommended Products
biological source
Serratia marcescens
Quality Level
recombinant
expressed in E. coli
Assay
≥90% (SDS-PAGE)
form
buffered aqueous glycerol solution
mol wt
30 kDa
concentration
≥250 units/μL
application(s)
research use
foreign activity
protease, essentially free
shipped in
wet ice
storage temp.
−20°C
Looking for similar products? Visit Product Comparison Guide
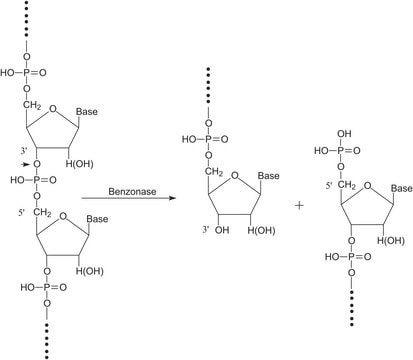
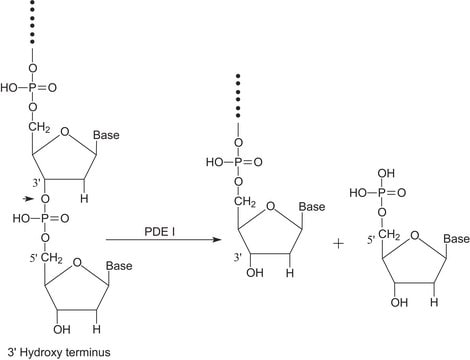
General description
Application
Biochem/physiol Actions
Features and Benefits
- Host DNA depletion in microbiome samples.
- Effective nucleic acid digestion in a variety of workflows.
- Viscosity reduction during protein extraction.
Unit Definition
Physical form
Legal Information
Storage Class Code
10 - Combustible liquids
WGK
WGK 1
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Personal Protective Equipment
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Articles
The field of proteomics is continually looking for new ways to investigate protein dynamics within complex biological samples. Recently, many researchers have begun to use RNA interference (RNAi) as a method of manipulating protein levels within their samples, but the ability to accurately determine these protein amounts remains a challenge. Fortunately, over the past decade, the field of proteomics has witnessed significant advances in the area of mass spectrometry. These advances, both in instrumentation and methodology, are providing researchers with sensitive assays for both identification and quantification of proteins within complex samples. This discussion will highlight some of these methodologies, namely the use of Multiple Reaction Monitoring (MRM) and Protein-AQUA.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service