KYSE-30
94072011, human esophagus, Polygonal
Synonym(s):
KYSE30 Cells
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
UNSPSC Code:
41106514
Recommended Products
Product Name
KYSE-30, 94072011
biological source
human esophagus
growth mode
Adherent
karyotype
Aneuploid
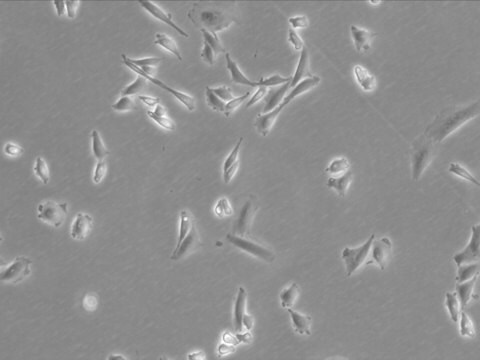
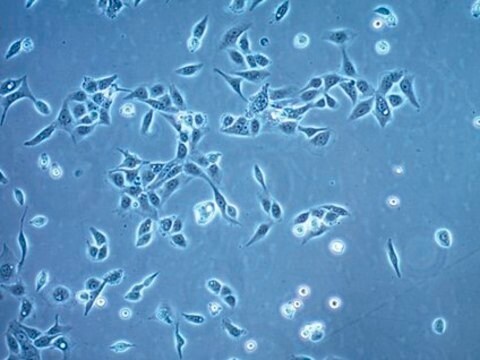
morphology
Polygonal
products
Not specified
receptors
Not specified
technique(s)
cell culture | mammalian: suitable
relevant disease(s)
cancer
shipped in
dry ice
storage temp.
−196°C
Cell Line Origin
Human Asian squamous cell carcinoma
Cell Line Description
KYSE-30 was established from the oesophageal cancer of an untreated 64 year old male. The tumour sample was taken from the mucosal surface of a well differentiated squamous cell carcinoma. The cell line KYSE-30 was established with the use of tumours initially transplanted to athymic mice. The cells are reported to have a doubling time of 20.8 hrs in the exponential growth phase. A p53 mutation at the splice acceptor site of intron 6 and a 12 fold amplification of c-erb B has been reported. KYSE-30 cells express a large number of epidermal growth factor receptors, 1.2x10,000,000 sites/cell.
Application
Study of human oesophageal cancer.
Culture Medium
RPMI 1640 + Ham′s F12 (1:1) + 2mM Glutamine + 2% Foetal Bovine Serum (FBS).
Subculture Routine
Split sub-confluent cultures (70-80%) 1:10 i.e. seeding at 1x10,000 cells/cm2 using 0.25% trypsin or trypsin/EDTA; 5% CO2; 37°C.
Other Notes
Additional freight & handling charges may be applicable for Asia-Pacific shipments. Please check with your local Customer Service representative for more information.
Choose from one of the most recent versions:
Certificates of Analysis (COA)
Lot/Batch Number
Sorry, we don't have COAs for this product available online at this time.
If you need assistance, please contact Customer Support.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service