GS70
GST P1-1, Recombinant Human
Synonym(s):
DFN7, FAEES3, GST3, GSTP, HEL-S-22, glutathione S-transferase pi
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
UNSPSC Code:
12352200
NACRES:
NA.26
Recommended Products
biological source
human
Quality Level
recombinant
expressed in E. coli
Assay
>95% (SDS-PAGE)
form
frozen liquid
specific activity
60.45 units/mg protein
mol wt
23 kDa
storage temp.
−70°C
Gene Information
human ... GSTP1(2950)
General description
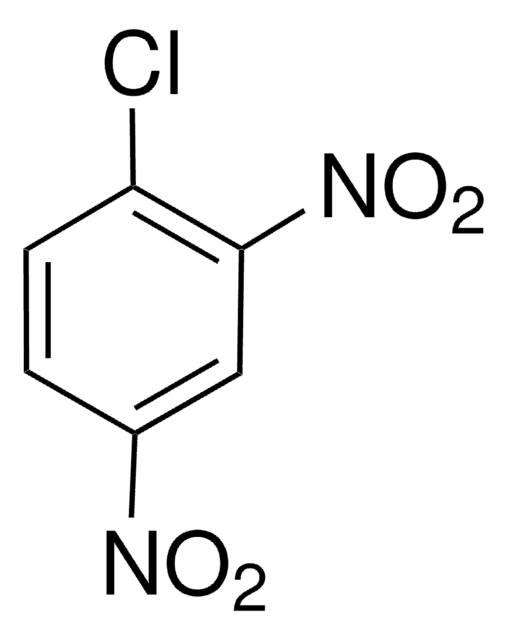
using spectrophotometric determination of 1-chloro-2,4-dinitrobenzene (CDNB) conjugation with reduced glutathione (1.0 mM) in 100 mM KPO4 (pH 6.5) at room temperature.
Biochem/physiol Actions
Glutathione S-transferase pi 1 (GSTP1) is an enzyme that in humans is encoded by the GSTP1 gene. Glutathione S-transferases (GSTs) are a family of enzymes that play an important role in detoxification by catalyzing the conjugation of many hydrophobic and electrophilic compounds with reduced glutathione. Based on their biochemical, immunologic, and structural properties, cytosolic and membrane-bound forms of glutathione S-transferase are encoded by two distinct supergene families. At present, eight distinct classes of the soluble cytoplasmic mammalian glutathione S-transferases have been identified: alpha, kappa, mu, omega, pi, sigma, theta and zeta. The GSTs are thought to function in xenobiotic metabolism and play a role in susceptibility to cancer, and other diseases.
This GST family member is a polymorphic gene encoding active, functionally different GSTP1 variant proteins that are thought to function in xenobiotic metabolism and play a role in susceptibility to cancer, and other diseases.
Storage and Stability
The enzyme should be used by the end-user customer within 1 year of receipt.
Storage Class Code
10 - Combustible liquids
WGK
WGK 1
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Choose from one of the most recent versions:
Certificates of Analysis (COA)
Lot/Batch Number
Don't see the Right Version?
If you require a particular version, you can look up a specific certificate by the Lot or Batch number.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
J Haluskova et al.
Bratislavske lekarske listy, 116(2), 79-82 (2015-02-11)
Prostate cancer (PCa) represents one of the most complicated human tumors and, like many others malignancies, arises from progressive genetic and epigenetic alterations. Among all recognized epigenetic alterations, aberrant DNA methylation (hypo- and hypermethylation) is the most important and the
Mohamad Nidal Khabaz et al.
Asian Pacific journal of cancer prevention : APJCP, 16(5), 1707-1713 (2015-03-17)
Eighty six cases of invasive ductal breast carcinomas were utilized to investigate GSTP1 polymorphisms in certain immunohistochemistry (IHC) subtypes of breast cancer with respect to ER, PR and HER2 expression. The frequency of wild allele homozygote, heterozygote and variant allele
Q F Li et al.
Genetics and molecular research : GMR, 14(2), 6762-6772 (2015-07-01)
Epigenetic silencing of the GSTP1 gene by promoter methylation has been associated with increased risk and shortened survival in patients with hepatocellular carcinoma (HCC). We therefore conducted a meta-analysis to obtain a more precise estimate of this association. By searching
Cheng-Fan Zhou et al.
Archives of dermatological research, 307(6), 505-513 (2015-06-06)
Numerous epidemiological studies have evaluated the association of Glutathione S-transferase P1 (GSTP1) Ile105Val polymorphism with the risk of skin cancer. However, the results remain inconclusive. To derive a more precise estimation of the association between the GSTP1 Ile105Val polymorphism and
Junhua Wu et al.
PloS one, 10(6), e0128280-e0128280 (2015-06-06)
Brick tea type fluorosis is a public health concern in the north-west area of China. The association between SNPs of genes influencing bone mass and fluorosis has attracted attention, but the association of SNPs with the risk of brick-tea type
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service