R1003
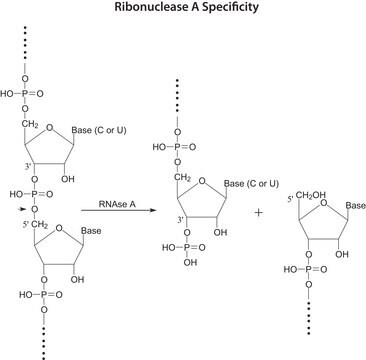
Ribonuclease T1 from Aspergillus oryzae
ammonium sulfate suspension, 300,000-600,000 units/mg protein
Synonym(s):
Guanyloribonuclease, Ribonucleate 3′-guanylo-oligonucleotidohydrolase
About This Item
Recommended Products
biological source
Aspergillus sp. (Aspergillus oryzae)
Quality Level
form
ammonium sulfate suspension
specific activity
300,000-600,000 units/mg protein
mol wt
11068 by amino acid sequence
technique(s)
cell based assay: suitable
suitability
suitable for separating native or denatured proteins, or nucleic acids
application(s)
cell analysis
storage temp.
2-8°C
Looking for similar products? Visit Product Comparison Guide
Application
Biochem/physiol Actions
Unit Definition
Physical form
Analysis Note
Storage Class Code
10 - Combustible liquids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Personal Protective Equipment
Regulatory Listings
Regulatory Listings are mainly provided for chemical products. Only limited information can be provided here for non-chemical products. No entry means none of the components are listed. It is the user’s obligation to ensure the safe and legal use of the product.
JAN Code
R1003-100KU:
R1003-VAR:
R1003-BULK:
R1003-500KU:
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Articles
Instructions for working with enzymes supplied as ammonium sulfate suspensions
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service