MBD0016
Inactivated Pseudomonas aeruginosa
Suitable for DNA extraction, PCR, sequencing, next generation sequencing, >10^8 bacteria/ml
About This Item
Recommended Products
Quality Level
form
liquid
concentration
>10^8 bacteria/ml
technique(s)
DNA extraction: suitable
DNA sequencing: suitable
PCR: suitable
shipped in
dry ice
storage temp.
−20°C
General description
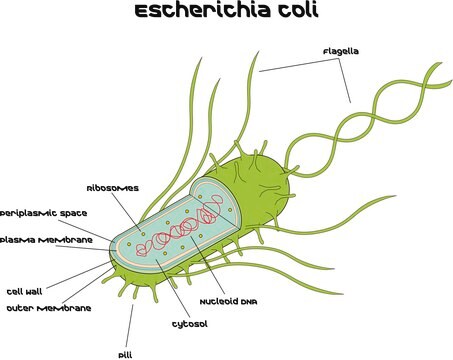
Pseudomonas aeruginosa is a gram negative aerobic to facultative anaerobe rod shaped bacteria which commonly inhabits soil and aqueous environments. In cases of a compromised immune system Pseudomonas aeruginosa can be an opportunistic pathogen. Its devastating effect is mostly known in Cystic Fibrosis (CF) patients. In addition, it often poses a major problem in burn wounds, chronic wounds, and in chronic obstructive pulmonary Disorder (COPD). P.aeruginosa forms biofilms on implanted biomaterials within hospital surface and water supplies. The international nosocomial infection control consortium reported that P. aeruginosa nosocomial infections have become a worldwide healthcare issue.
Read here how to use our standards to ensure data integrity for your microbiome research.
Application
Suitable for Quantitative standard for PCR, Sequencing and NGS
Features and Benefits
- Individual microbial standard for microbiomics and meta-genomics workflow
- Suitable standard for PCR, sequencing and NGS
- Improve Bioinformatics analyses
- Increases reproducibility
- Compare results lab to lab
Physical form
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 2
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Choose from one of the most recent versions:
Certificates of Analysis (COA)
Don't see the Right Version?
If you require a particular version, you can look up a specific certificate by the Lot or Batch number.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Articles
An overview of human microbiome research, workflow challenges, sequencing, library production, data analysis, and available microbiome reagents to support your research.
The use of standards is critical to the integrity of metagenomics research. Learn how DNA standards for bacteria, fungi, and viruses are applied to studying the microbiome. Choose standards for E. coli and other key species, as well as mixed community standards.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service