10107921001
Roche
Nuclease S7
Micrococcal nuclease, from Staphylococcus aureus
Synonym(s):
s7 nuclease
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
UNSPSC Code:
12352204
Recommended Products
biological source
Staphylococcus aureus
Quality Level
form
lyophilized
specific activity
~15,000 U/mg
packaging
pkg of 15,000 U
manufacturer/tradename
Roche
optimum pH
7.5
shipped in
wet ice
Application
- Removal of endogenous RNA from in vitro translation systems
- RNA sequencing experiments
Nuclease S7 has been used in ribonuclease selection process for ribosome profiling.
Biochem/physiol Actions
Nuclease S7 is an endonuclease that mainly cleaves single-stranded substrates. It also cleaves double-stranded DNA or RNA. It cleaves the 5′-phosphodiester bonds of RNA and DNA to yield 3′-mononucleotides and oligonucleotides. Activity of the enzyme is dependent on Ca2+ and can be completely inactivated by the addition of EDTA. Nuclease S7 cleavage is highly specific to AT rich region than GC on the DNA.
Unit Definition
One unit releases a sufficient amount of acid-soluble oligonucleotides to produce an increase of 1.0 at A260.
Storage and Stability
Store unopened reagent at 2-8 °C; when diluted to 1 mg/mL, store at -15 to -25 °C.
Other Notes
For life science research only. Not for use in diagnostic procedures.
Signal Word
Danger
Hazard Statements
Precautionary Statements
Hazard Classifications
Eye Irrit. 2 - Resp. Sens. 1 - Skin Irrit. 2 - Skin Sens. 1 - STOT SE 3
Target Organs
Respiratory system
Storage Class Code
11 - Combustible Solids
WGK
WGK 1
Flash Point(F)
does not flash
Flash Point(C)
does not flash
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Reimo Zoschke et al.
Proceedings of the National Academy of Sciences of the United States of America, 112(13), E1678-E1687 (2015-03-17)
Chloroplast genomes encode ∼ 37 proteins that integrate into the thylakoid membrane. The mechanisms that target these proteins to the membrane are largely unexplored. We used ribosome profiling to provide a comprehensive, high-resolution map of ribosome positions on chloroplast mRNAs
Serap Erkek et al.
Nature structural & molecular biology, 20(7), 868-875 (2013-06-19)
In mammalian spermatozoa, most but not all of the genome is densely packaged by protamines. Here we reveal the molecular logic underlying the retention of nucleosomes in mouse spermatozoa, which contain only 1% residual histones. We observe high enrichment throughout
Ribonuclease selection for ribosome profiling.
Gerashchenko M V and Gladyshev V N
Nucleic Acids Research, 45(2), e6-e6 (2016)
Maxim V Gerashchenko et al.
Nucleic acids research, 49(2), e9-e9 (2020-12-03)
There has been a surge of interest towards targeting protein synthesis to treat diseases and extend lifespan. Despite the progress, few options are available to assess translation in live animals, as their complexity limits the repertoire of experimental tools to
Sarah O'Keefe et al.
bioRxiv : the preprint server for biology (2020-12-04)
In order to produce proteins essential for their propagation, many pathogenic human viruses, including SARS-CoV-2 the causative agent of COVID-19 respiratory disease, commandeer host biosynthetic machineries and mechanisms. Three major structural proteins, the spike, envelope and membrane proteins, are amongst
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service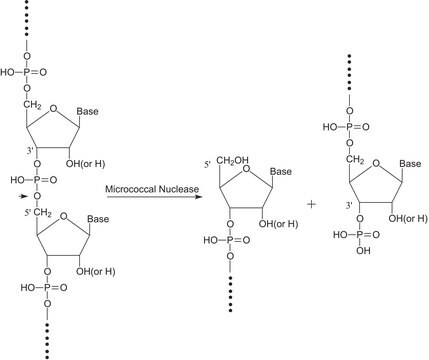







![Guanosine 5′-[β,γ-imido]triphosphate trisodium salt hydrate ≥85% (HPLC), powder](/deepweb/assets/sigmaaldrich/product/structures/204/494/05808804-1ca7-44bf-b6c5-d4934dc7cb85/640/05808804-1ca7-44bf-b6c5-d4934dc7cb85.png)