03353591910
Roche
DIG Gel Shift Kit, 2nd generation
storage temp.:-20°C (-15°C to -25°C)
About This Item
Recommended Products
description
New formulation containing Terminal Transferase, recombinant.
Quality Level
manufacturer/tradename
Roche
greener alternative product characteristics
Designing Safer Chemicals
Learn more about the Principles of Green Chemistry.
sustainability
Greener Alternative Product
greener alternative category
, Aligned
shipped in
dry ice
storage temp.
−20°C (−15°C to −25°C)
General description
DIG Gel Shift Kit employs recombinant terminal transferase and digoxigenin (DIG)-11- dideoxyuridine triphosphate (ddUTP) makes the labeling reaction very flexible. It can be used to label the 3′ ends of any oligonucleotide (whether it has a 5′- overhanging end, a 3′-overhanging end, or blunt ends). Both single- and double-stranded DNA can be labeled.
Application
Features and Benefits
- Convenient, since the technique does not require special equipment or technology.
- Reproducible, since DIG-labeled probes are stable indefinitely.
- Safe, because the assay is nonradioactive.
- Reliable, because the kit provides DIG-labeled control oligonucleotides to establish that the assay is working.
- Function-tested with the controls provided in the kit (See "Quality").
- Sensitive, since the kit can detect as little as 20fmol of the control oligonucleotide (after it is labeled according to the kit protocol).
Packaging
Physical form
Preparation Note
Sample
Amount: 3.85pmol or 100ng
Type: Oligonucleotides with 5′-overhanging ends, 3′-overhanging ends, or blunt ends; single- or double-stranded DNA fragments (30 - 200bp)
Note: Ideally, fragments to be labeled should be between 30 and 100bp.
Time required
Oligonucleotide annealing and labeling: 10minutes
Formation of oligonucleotide-protein complexes: 25minutes
Electrophoresis: 1 hour to overnight, depending on electrophoresis system
Blotting: 1 - 2 hours
Immunological detection: 2hours
Exposure to X-ray film or imager: 15 - 40minutes
Other Notes
Kit Components Only
- Labeling Buffer 5x concentrated
- CoCl<SUB>2</SUB> Solution 25 mM
- DIG-ddUTP Solution, 1 mM Digoxigenin-11-ddUTP
- Recombinant Terminal Transferase 400 U/μl
- Binding Buffer 5x concentrated
- Control Oligonucletide ds 39mer, unlabeled 0.1 μg/μl, 3.85 pmol/μl
- DIG-labeled Control Oligonucleotide ds 39mer 0.4 ng/μl, 15.54 fmol/μl
- Control Factor Oct2A 25-75 ng/μl
- Poly [d(I-C)] 0.1 μg/μl
- Poly [d(A-T)] 0.1 μg/μl
- Poly L-lysine 0.1 μg/μl
- Loading Buffer without bromophenol blue
- Loading Buffer with bromophenol blue
- Anti-Digoxigenin-AP 750 U/ml
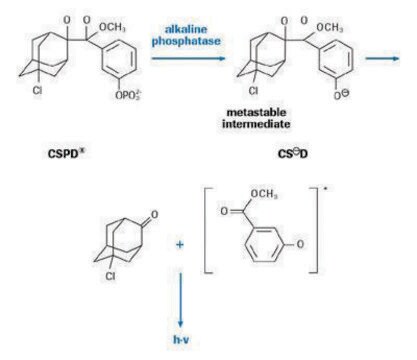
- CSPD 10 mg/ml
- Blocking Reagent
Signal Word
Danger
Hazard Statements
Precautionary Statements
Hazard Classifications
Acute Tox. 4 Inhalation - Acute Tox. 4 Oral - Aquatic Chronic 2 - Carc. 1B Inhalation - Repr. 1B
Storage Class Code
6.1D - Non-combustible, acute toxic Cat.3 / toxic hazardous materials or hazardous materials causing chronic effects
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Articles
Digoxigenin (DIG) labeling methods and kits for DNA and RNA DIG probes, random primed DNA labeling, nick translation labeling, 5’ and 3’ oligonucleotide end-labeling.
Protocols
DIG Gel Shift Kit 2nd Generation Protocol & Troubleshooting
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service

![Poly[d(I-C)] lyophilized, pkg of 10 U (10108812001 [A<sub>260</sub> units]), pkg of 50 U (11219847001 [A<sub>260</sub> units])](/deepweb/assets/sigmaaldrich/product/images/352/091/ef743cea-ccd8-44f1-8f3b-dec5a1e4f5d1/640/ef743cea-ccd8-44f1-8f3b-dec5a1e4f5d1.jpg)