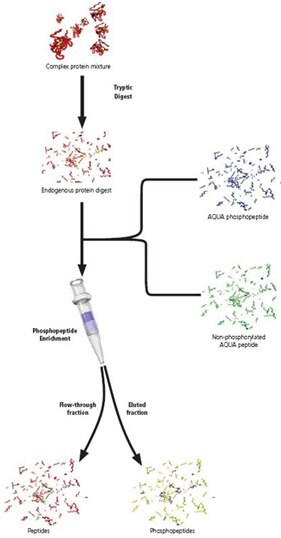
In June 2003, Dr. Steve Gygi and his team presented an innovative strategy, Protein AQUA™, enabling absolute protein quantitation using stable isotope labeled peptides and HPLC-MS. By applying a common principle, the use of a labeled molecule as an internal standard, to protein analysis, Gygi’s team has advanced the abilities of protein researchers to study complex biological samples quantitatively and has provided a valuable new tool for Proteomics.
An AQUA™ Peptide is simply a synthetic tryptic peptide corresponding to a peptide of interest. Each AQUA™ peptide incorporates one stable isotope labeled amino acid, creating a slight increase (6-10 daltons) in molecular weight. When mixed, the native peptide and the synthetic AQUA™ Peptide elute together chromatographically, migrate together electrophoreticly, and ionize with the same intensity. However, by mass spectrometry, the native peptide and the synthetic AQUA™ Peptide are easily distinguished.
In a typical AQUA™ experiment, a known amount of AQUA™ Peptide is added to a biological protein sample. The sample is then digested and analyzed by HPLC-MS. Extracted ion chromatograms are generated for the native peptide and the synthetic AQUA™ Peptide internal standard. Using peak ratios, the quantity of native peptide is calculated.
Protein-AQUA™ is a powerful, enabling technology, the limits of which are only now being explored. For proteomics researchers, it facilitates focused, quantitative studies of not only specific protein expression but specific amino acid modification as well.
Verify Gene Silencing
Verification of gene silencing represents a novel and powerful application of the AQUA technique. Recently, we demonstrated that AQUA quantitation is an effective method for measuring expression of silenced genes and verifying knockdown success (Figure 1). AQUA quantitation offers many advantages over traditional methods for measuring expression of silenced genes:
- AQUA sensitivity is limited only by the sensitivity of the mass spectrometer, enabling researchers to study gene silencing in low abundance proteins, or to detect very low expression of silenced genes. This represents a marked advantage over traditional methods of Western blotting and ELISA.
- An AQUA™ peptide can be generated using only an amino acid sequence, even when no native peptide has been isolated or when no corresponding antibody exists. For this reason, AQUA can be used to measure any silenced gene while alternative methods, such as Western blotting, can not.
- AQUA quantitation measures protein (and corresponding peptide) expression. This provides a more accurate correlation to silencing efficiency when compared to quantitative PCR or Northern blotting.
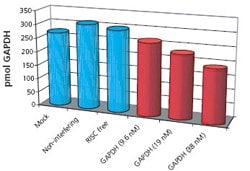
Figure 1: HeLa cultures transfected with siRNA for Human GAPDH (9.6 nM, 19 nM, and 38 nM), RISC free (non-functional, non-interfering), two non-interfering sequences, and a non-interfering pool. Additionally, four mock transfections were performed. Transfected cells were incubated 48 additional hours. Proteins were extracted, precipitated, quantitatively spiked with AQUA™ peptides, tryptically digested, and analyzed by reversed phase LC-MS. Restricted ion current chromatograms for native and AQUA™ peptides from GAPDH were generated; based on peak area ratios, GAPDH concentrations were calculated.
Increase the sensitivity of Protein-AQUA™ by depleting high-abundance proteins
The study of the plasma proteome has historically been fundamentally limited by its vast dynamic range (10 orders of magnitude). Low-abundance proteins, carrying great diagnostic potential, are often obscured by the presence of high-abundance serum proteins. Recently, Jeffrey Porter and his team demonstrated an effective strategy for both identifying and quantitating numerous lower-abundance proteins in human serum by combining ProteoPrep® 20 Plasma Immunodepletion Kit and the Protein AQUA™ technology. This combination of technologies may provide a targeted approach to the discovery of proteins holding biological and diagnostic significance (Figure 1).
Learn more about this experiment when you download the poster Absolute Quantification of the Lower Abundance Proteome Through Immunoaffinity Depletion of the Twenty Most Abundant Proteins in Human Serum
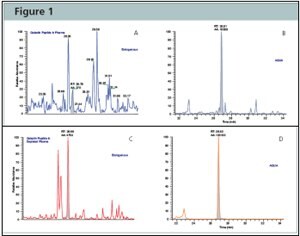
Figure 2:Protein-AQUA™ Analysis of Gelsolin in Whole Plasma and ProteoPrep® 20 Depleted Samples. SRM Analysis of the endogenous gelsolin peptide, GASQAGAPQGR, results in a complex chromatogram in whole plasma (Figure A) with poor resolution and low signal-to-noise. When analyzed in depleted plasma (Figure C), the endogenous gelsolin peptide is easily separated and identified from interfering species and its concentration readily calculated by comparison to the AQUA™ peptide internal standard (Figures B and D).
Enhance sensitivity of Protein-AQUA™ of Phosphopeptides through enrichment with a Gallium-IMAC Spin Column
The analysis of phosphorylated peptides by mass spectrometry has often been limited by their intrinsically low abundance and poor ionization. Recently, John Dapron and his team demonstrated an effective strategy to both enhance phosphopeptide MS signal levels and quantitate specific phosphopeptides using Protein-AQUA™. Using an Gallium Immobilized Metal Affinity Chromatography (IMAC) based microspin column and phosphorylated AQUA™ peptides, Dapron was able to easily enrich, identify, and quantitate specific phosphopeptides under varying biological conditions. In the course of his experiments, Dapron examined the sensitivity levels achieved by coupling the Gallium IMAC microspin column with Protein-AQUA™. He spiked a control mixture of phosphorylated and non-phosphorylated AQUA™ peptides (at 39 ng, 3.9 ng, and 390 pg levels) into an E. coli protein digest, enriched using the microspin column, and analyzed the eluted fractions by LC-MS. The extracted ion chromatograms indicate the 39 ng and 3.9 ng levels were easily observed. Using single reaction monitoring (SRM), the 390 pg phosphopeptide was also easily observed.
如要继续阅读,请登录或创建帐户。
暂无帐户?