EHU113021
MISSION® esiRNA
targeting human YAP1
About This Item
Produits recommandés
Description
Powered by Eupheria Biotech
Niveau de qualité
Gamme de produits
MISSION®
Forme
lyophilized powder
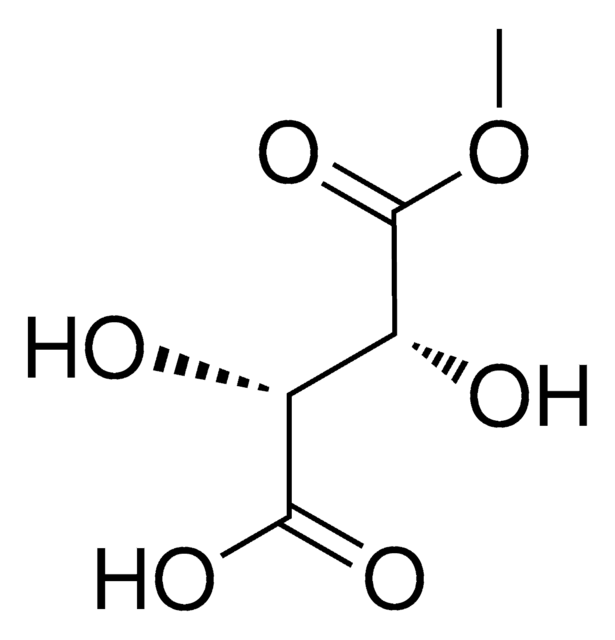
Séquence cible d'ADNc esiRNA
GTCTTCTCCCGGGATGTCTCAGGAATTGAGAACAATGACGACCAATAGCTCAGATCCTTTCCTTAACAGTGGCACCTATCACTCTCGAGATGAGAGTACAGACAGTGGACTAAGCATGAGCAGCTACAGTGTCCCTCGAACCCCAGATGACTTCCTGAACAGTGTGGATGAGATGGATACAGGTGATACTATCAACCAAAGCACCCTGCCCTCACAGCAGAACCGTTTCCCAGACTACCTTGAAGCCATTCCTGGGACAAATGTGGACCTTGGAACACTGGAAGGAGATGGAATGAACATAGAAGGAGAGGAGCTGATGCCAAGTCTGCAGGAAGCTTTGAGTTCTGACATCCTTAATGACATGGAGTCTGTTTTGGCTGCCACCAAGCTAGATAAAGAAAGCTTTCTTACATGGTTATAGAGCCCTCAGGCAGACTGAATTCTAAATCTGTGAAGGATCTAAGGAGACACATGCACCGGAAATT
Numéro d'accès Ensembl | humain
Numéro d'accès NCBI
Conditions d'expédition
ambient
Température de stockage
−20°C
Informations sur le gène
human ... YAP1(10413) , YAP1(10413)
Description générale
For additional details as well as to view all available esiRNA options, please visit SigmaAldrich.com/esiRNA.
Informations légales
Code de la classe de stockage
10 - Combustible liquids
Point d'éclair (°F)
Not applicable
Point d'éclair (°C)
Not applicable
Faites votre choix parmi les versions les plus récentes :
Déjà en possession de ce produit ?
Retrouvez la documentation relative aux produits que vous avez récemment achetés dans la Bibliothèque de documents.
Global Trade Item Number
| Référence | GTIN |
|---|---|
| EHU113021-50UG | 4061831374858 |
| EHU113021-20UG | 4061831358537 |
Notre équipe de scientifiques dispose d'une expérience dans tous les secteurs de la recherche, notamment en sciences de la vie, science des matériaux, synthèse chimique, chromatographie, analyse et dans de nombreux autres domaines..
Contacter notre Service technique