11062590001
Roche
DNA Molecular Weight Marker VI
pkg of 50 μg (in 200 μl), solution
Synonyme(s) :
DNA marker
Se connecterpour consulter vos tarifs contractuels et ceux de votre entreprise/organisme
About This Item
Code UNSPSC :
41105335
Produits recommandés
Forme
solution
Niveau de qualité
Conditionnement
pkg of 50 μg (in 200 μl)
Fabricant/nom de marque
Roche
Concentration
250 μg/mL
Couleur
colorless
Solubilité
water: miscible
Température de stockage
−20°C
Description générale
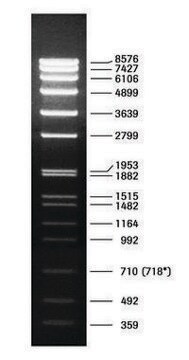
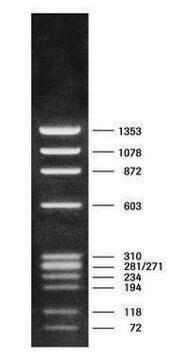
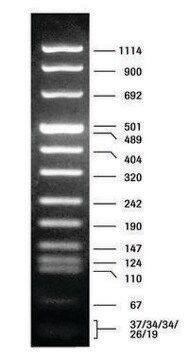
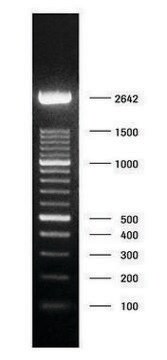
DNA Molecular Weight Marker VI is a fragment mixture prepared by cleavage of pBR328 DNA with restriction endonucleases Bgl I and pBR328 DNA digested with Hinf I. It has a size range of 0.15 to 2.1kbp.
The product is also available as DNA Molecular Weight Marker VI, Digoxigenin-labeled.
The product is also available as DNA Molecular Weight Marker VI, Digoxigenin-labeled.
Spécificité
Specificity: Product has 5′-phosphorylated ends (restriction fragments).
Restriction site: Claevage sites and corresponding fragment lengths of pBR328:
- Bgl I cut sites at: 935, 1169, 2399, 4575, 4809 => creating following fragment sizes, respectively: 234 bp, 1230 bp, 2176 bp, 234 bp, 1033 bp.
- Hinf I cut sites at: 632, 852, 1006, 1304, 1757, 2274, 4040, 4434, 4732, 4886. =>
creating following fragment sizes, respectively: 220 bp, 154 bp, 298 bp, 453 bp, 517 bp,
1766 bp, 394 bp, 298 bp, 154 bp, 653 bp
Restriction site: Claevage sites and corresponding fragment lengths of pBR328:
- Bgl I cut sites at: 935, 1169, 2399, 4575, 4809 => creating following fragment sizes, respectively: 234 bp, 1230 bp, 2176 bp, 234 bp, 1033 bp.
- Hinf I cut sites at: 632, 852, 1006, 1304, 1757, 2274, 4040, 4434, 4732, 4886. =>
creating following fragment sizes, respectively: 220 bp, 154 bp, 298 bp, 453 bp, 517 bp,
1766 bp, 394 bp, 298 bp, 154 bp, 653 bp
Application
DNA Molecular Weight Marker VI is used for the size determination of DNA in agarose gels. It simplifies accurate molecular weight determination of double-stranded DNA fragments generated by:
- Restriction digests
- PCR
- RT-PCR
Séquence
As determined by computer analysis of the pBR328 sequence, the mixture contains 15 DNA fragments with the following base pair lengths (1 base pair = 660 daltons) occuring once: 220, 394, 453, 517, 653, 1033, 1230, 1766, 2176 and fragments 154,234, and 298 occuring twice as a result of the digestion of pBR328 with both Bgl I and Hinf I.
Note: Fragment lengths are derived from computer analysis of the DNA sequence. Depending on the size range of the marker, the smallest fragments will only be visible on overloaded gels.
Note: Fragment lengths are derived from computer analysis of the DNA sequence. Depending on the size range of the marker, the smallest fragments will only be visible on overloaded gels.
Définition de l'unité
50 μg = 1A260 unit
Forme physique
Ready-to-use solution, 250μg/ml, in TE buffer (10mM Tris-HCl, 1mM EDTA, pH 8.0).
Notes préparatoires
Storage conditions (working solution): 2 to 8 °C
Once thawed we recommend further storage at 2 to 8 °C.
Once thawed we recommend further storage at 2 to 8 °C.
Autres remarques
For life science research only. Not for use in diagnostic procedures.
Code de la classe de stockage
12 - Non Combustible Liquids
Classe de danger pour l'eau (WGK)
nwg
Point d'éclair (°F)
does not flash
Point d'éclair (°C)
does not flash
Certificats d'analyse (COA)
Recherchez un Certificats d'analyse (COA) en saisissant le numéro de lot du produit. Les numéros de lot figurent sur l'étiquette du produit après les mots "Lot" ou "Batch".
Déjà en possession de ce produit ?
Retrouvez la documentation relative aux produits que vous avez récemment achetés dans la Bibliothèque de documents.
Les clients ont également consulté
G Meulemans et al.
Avian pathology : journal of the W.V.P.A, 30(6), 655-660 (2001-12-01)
A polymerase chain reaction combined with restriction enzyme analysis was developed for detection and differentiation of all 12 fowl adenovirus (FAdV) serotypes representing the five fowl adenovirus (A to E) species. For primer design, the published sequences of the hexon
Qualitative PCR?ELISA protocol for the detection and
typing of viral genomes
typing of viral genomes
Monica Musiani
Nature Protocols (2007)
R Ferretti et al.
Applied and environmental microbiology, 67(2), 977-978 (2001-02-07)
A PCR-based method for the detection of Salmonella spp. in food was developed. The method, set up on typical salami from the Italian region of Marche, is sensitive and specific and shows excellent correlation with the conventional method of reference
Notre équipe de scientifiques dispose d'une expérience dans tous les secteurs de la recherche, notamment en sciences de la vie, science des matériaux, synthèse chimique, chromatographie, analyse et dans de nombreux autres domaines..
Contacter notre Service technique