SAE0045
HRV-3C Protease.
N-Terminal His tagged recombinant protein, aqueous solution, 0.8-1.2 mg/mL
Synonym(s):
Human Rhinovirus 3C Protease, Levlfqgp site protease, PreScission Protease
About This Item
Recommended Products
biological source
human (human Rhinovirus Type 14)
Quality Level
Assay
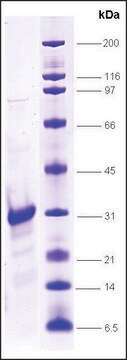
≥90% (SDS-PAGE)
form
aqueous solution
specific activity
≥5000 U/mg
mol wt
21 kDa
concentration
0.8-1.2 mg/mL
technique(s)
protein purification: suitable
suitability
suitable for protein modification
application(s)
life science and biopharma
shipped in
dry ice
storage temp.
−20°C
General description
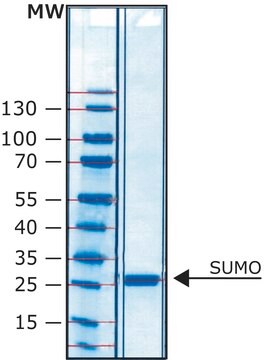
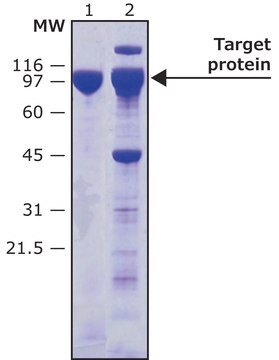
Proteolytic cleavage occurs between the Gln and Gly residues.
HRV-3C protease is a useful tool to cleave recombinant proteins that are expressed as a fusion protein with this sequence, between the carrier domain and the protein of interest.
This recombinant version contains a six-histidine tag and can be easily removed by IMAC chromatography.
Application
Unit Definition
Physical form
Storage Class Code
10 - Combustible liquids
WGK
WGK 2
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service