10976776001
Roche
Nick Translation Kit
sufficient for 50 labeling reactions, kit of 1 (7 components), suitable for FISH
Sinónimos:
nick translation
Iniciar sesiónpara Ver la Fijación de precios por contrato y de la organización
About This Item
Código UNSPSC:
41105500
Productos recomendados
uso
sufficient for 50 labeling reactions
Nivel de calidad
envase
kit of 1 (7 components)
fabricante / nombre comercial
Roche
técnicas
FISH: suitable
temp. de almacenamiento
−20°C
Categorías relacionadas
Descripción general
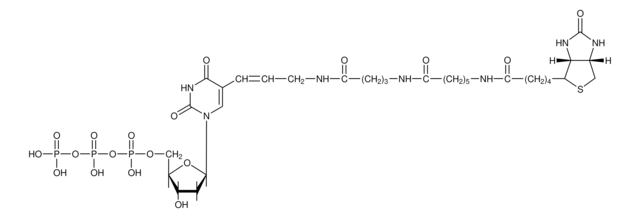
Kit for radioactive or nonradioactive labeling of DNA by the nick translation method. This method utilizes a combination of DNase and DNA Polymerase to nick one strand of the DNA helix, then incorporates labeled nucleotides as the polymerase examines, or "proofreads" the nicked site.
Especificidad
Heat inactivation: Stop the reaction by adding 1μl 0.5 M EDTA (pH 8.0) and/or by heating to 65 °C for 10 minutes.
Aplicación
Nick translation kit has been used in FISH (fluorescence in situ hybridization) based labeling of tick and bacterial artificial chromosome. It has been used in labeling the probe (genomic DNA) in order to obtain GISH (genomic in situ hybridization) signals.
The kit is used for labeling of DNA with radioactive or modified dNTPs. Probes labeled by nick translation are used in many different hybridization techniques. These include use as probes in:
- Screening gene banks by colony- or plaque hybridization
- DNA or RNA transfer hybridizations
- In situ hybridization
- Reassociation kinetic studies
Envase
1 kit containing 7 components.
Especificaciones
Assay Time: 45minutes
Specific Activity: The level of specific labeling and the incorporation rate are dependent on the ratio of substrate DNA to labeled deoxyribonucleoside triphosphate, e.g., the kinetics and labeling levels obtained are identical in assays containing 0.1μg DNA and 20μCi dXTP or 0.5μg DNA and 100μCi dXTP.
The standard assay will give a specific activity of 3 x 108 dpm/μg, corresponding to 65% incorporation with different substrate DNAs (e.g., pBR322, λDNA, DNA fragments) in 35minutes.
Sample Materials
Specific Activity: The level of specific labeling and the incorporation rate are dependent on the ratio of substrate DNA to labeled deoxyribonucleoside triphosphate, e.g., the kinetics and labeling levels obtained are identical in assays containing 0.1μg DNA and 20μCi dXTP or 0.5μg DNA and 100μCi dXTP.
The standard assay will give a specific activity of 3 x 108 dpm/μg, corresponding to 65% incorporation with different substrate DNAs (e.g., pBR322, λDNA, DNA fragments) in 35minutes.
Sample Materials
- Supercoiled and linearized plasmid DNA
- Supercoiled and linearized cosmid DNA
- BAC DNA or human genomic DNA
- Purified PCR products
Principio
The nick translation method is based on the ability of DNase I to introduce randomly distributed nicks into DNA at low enzyme concentrations in the presence of Mg2+. E. coli DNA polymerase I synthesizes DNA complementary to the intact strand in a 5′→3′ direction using the 3′-OH termini of the nick as a primer. The 5′→3′ exonucleolytic activity of DNA polymerase I simultaneously removes nucleotides in the direction of synthesis. The polymerase activity sequentially replaces the removed nucleotides with isotope-labeled or hapten-labeled deoxyribonucleoside triphosphates. At low temperature (+15°C), the unlabeled DNA in the reaction is thus replaced by newly synthesized labeled DNA.
Nota de preparación
Working concentration: The recommended working concentration with T7, T3 or SP6 RNA Polymerases is 1 mM.
Working solution: DNA
The DNA must be in low-salt concentration solution.
dATP, dGTP, dTTP
Working solution: DNA
The DNA must be in low-salt concentration solution.
dATP, dGTP, dTTP
- If the same labeled deoxyribonucleoside triphosphate is used repeatedly, we recommend the preparation of a mixture of equal parts of the other three triphosphates for convenience.
- Prepare the dATP, dGTP, dTTP mixture by making a 1:1:1 mixture of solution 2, solution 4, and solution 5.
Otras notas
For life science research only. Not for use in diagnostic procedures.
Solo componentes del kit
Referencia del producto
Descripción
- Control DNA pBR322, 20 μl 50 µg/ml
- dATP, 50 μl 0.4 mM
- dCTP, 50 μl 0.4 mM
- dGTP, 50 μl 0.4 mM
- dTTP, 50 μl 0.4 mM
- Nick Translation Buffer, 100 μl 10x concentrated
- Enzyme Mixture, 100 μl, DNA Polymerase I and DNase I in 50% glycerol (v/v)
Código de clase de almacenamiento
12 - Non Combustible Liquids
Clase de riesgo para el agua (WGK)
WGK 1
Punto de inflamabilidad (°F)
does not flash
Punto de inflamabilidad (°C)
does not flash
Certificados de análisis (COA)
Busque Certificados de análisis (COA) introduciendo el número de lote del producto. Los números de lote se encuentran en la etiqueta del producto después de las palabras «Lot» o «Batch»
¿Ya tiene este producto?
Encuentre la documentación para los productos que ha comprado recientemente en la Biblioteca de documentos.
Los clientes también vieron
Amy Sebeson et al.
PloS one, 10(5), e0127214-e0127214 (2015-05-21)
The binding sequence for any transcription factor can be found millions of times within a genome, yet only a small fraction of these sequences encode functional transcription factor binding sites. One of the reasons for this dichotomy is that many
A methylation status analysis of the apomixis-specific region in Paspalum spp. suggests an epigenetic control of parthenogenesis.
Podio M, et al.
Journal of Experimental Botany, 65(22), 6411-6424 (2014)
David U Gorkin et al.
Genome biology, 20(1), 255-255 (2019-11-30)
The 3-dimensional (3D) conformation of chromatin inside the nucleus is integral to a variety of nuclear processes including transcriptional regulation, DNA replication, and DNA damage repair. Aberrations in 3D chromatin conformation have been implicated in developmental abnormalities and cancer. Despite
Aiko Iwata-Otsubo et al.
Current biology : CB, 27(15), 2365-2373 (2017-08-02)
Female meiosis provides an opportunity for selfish genetic elements to violate Mendel's law of segregation by increasing the chance of segregating to the egg [1]. Centromeres and other repetitive sequences can drive in meiosis by cheating the segregation process [2]
Anna Zaleśna et al.
Comparative cytogenetics, 11(2), 249-266 (2017-09-19)
We studied water frogs from a complex composed of two species: Pelophylax lessonae (Camerano, 1882) (genome LL, 2n = 26) and P. ridibundus (Pallas, 1771) (RR, 2 = 26), and their natural hybrid P. esculentus (Fitzinger, 1843) of various ploidy
Nuestro equipo de científicos tiene experiencia en todas las áreas de investigación: Ciencias de la vida, Ciencia de los materiales, Síntesis química, Cromatografía, Analítica y muchas otras.
Póngase en contacto con el Servicio técnico