MBD0001
Microbial DNA standard from Akkermansia muciniphila
Suitable for PCR, sequencing and NGS, 10 ng/μL
About This Item
Recommended Products
Quality Level
form
liquid
concentration
10 ng/μL
technique(s)
DNA extraction: suitable
DNA sequencing: suitable
PCR: suitable
shipped in
ambient
storage temp.
−20°C
Related Categories
General description
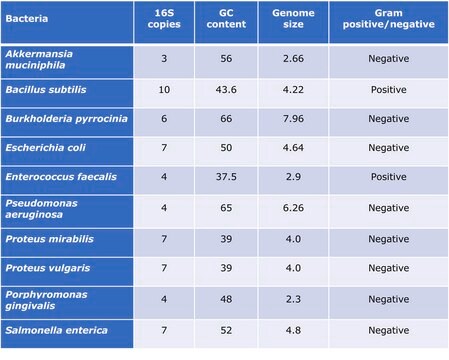
Akkermansia muciniphila is a gram negative, anaerobic, non motile, non spore forming and oval shaped bacterium. A. muciniphila inhabits the gastrointestinal tracts of more than 90% of adults and constitutes 1 to 4% of the fecal microbiota.2 A. muciniphila level was suggested to be a biomarker for a healthy intestin. 3
A. muciniphila degrades mucus and utilizes it as a carbon/nitrogen source. Consequently, the host produces additional mucus while the bacterium produces oligosaccharides and Short Chain Fatty Acids (SCFAs) that can be utilized by the host and trigger the immune system. An additional protective effect of the SCFA is stimulation of mucus-associated microbiota growth, that serves as a barrier against penetration of pathogens to intestinal cells. 2,4
Read here how to use our standards to ensure data integrity for your microbiome research.
Application
Features and Benefits
- Individual microbial standard for microbiomics and meta-genomics workflow
- Suitable standard for PCR, sequencing and NGS
- Improve Bioinformatics analyses
- Increases reproducibility
- Compare results lab to lab
Physical form
Other Notes
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 1
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Choose from one of the most recent versions:
Certificates of Analysis (COA)
Don't see the Right Version?
If you require a particular version, you can look up a specific certificate by the Lot or Batch number.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Articles
An overview of human microbiome research, workflow challenges, sequencing, library production, data analysis, and available microbiome reagents to support your research.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service