R6750
Ribonucleic acid from baker′s yeast (S. cerevisiae)
Synonym(s):
RNA
Sign Into View Organizational & Contract Pricing
All Photos(4)
About This Item
Recommended Products
Quality Level
storage temp.
−20°C
Looking for similar products? Visit Product Comparison Guide
General description
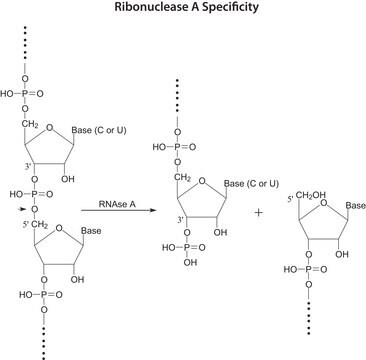
Ribonucleic Acid (RNA) from baker’s yeast (S. cerevisiae) is the substrate for RNase enzyme. The isolation of RNA from yeast is a complicated process and involves heating and freezing cycles of cells in the presence of phenyl and detergents (SDS).
Application
Ribonucleic Acid (RNA) from baker’s yeast (S. cerevisiae) is suitable for applications such as northern blot hybridization, reverse transcriptase-polymerase chain reaction (RTPCR), and cDNA construction.
Preparation Note
Prepared by a modification of Crestfield, A.M., et al., J. Biol. Chem., 216, 185 (1955).
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Personal Protective Equipment
dust mask type N95 (US), Eyeshields, Gloves
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Lukas Bossaller et al.
Journal of immunology (Baltimore, Md. : 1950), 197(4), 1044-1053 (2016-06-30)
Systemic lupus erythematosus (SLE) is a chronic, life-threatening autoimmune disorder, leading to multiple organ pathologies and kidney destruction. Analyses of numerous murine models of spontaneous SLE have revealed a critical role for endosomal TLRs in the production of autoantibodies and
Hidekazu Iioka et al.
Nucleic acids research, 39(8), e53-e53 (2011-02-09)
The diverse localization of transcripts in cells suggests that there are many specific RNA-protein interactions that have yet to be identified. Progress has been limited, however, by the lack of a robust method to detect and isolate the RNA-binding proteins.
Michael R Green et al.
Cold Spring Harbor protocols, 2021(12) (2021-12-03)
Isolation of RNA from yeast is complicated by the need to first break the thick, rigid cell wall. The protocol provided here uses a cycle of heating and freezing of cells in the presence of phenol and the detergent sodium
Stephanie C Bannister et al.
Biology of reproduction, 81(1), 165-176 (2009-04-10)
MicroRNAs are a highly conserved class of small RNAs that function in a sequence-specific manner to posttranscriptionally regulate gene expression. Tissue-specific miRNA expression studies have discovered numerous functions for miRNAs in various aspects of embryogenesis, but a role for miRNAs
Hilal Kazan et al.
Nucleic acids research, 41(Web Server issue), W180-W186 (2013-06-12)
RBPmotif web server (http://www.rnamotif.org) implements tools to identify binding preferences of RNA-binding proteins (RBPs). Given a set of sequences that are known to be bound and unbound by the RBP of interest, RBPmotif provides two types of analysis: (i) de
Protocols
This procedure may be used for determination of Ribonuclease A (RNase A) activity.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service