10786357001
Roche
Ribonuclease H (RNase H)
from Escherichia coli H 560 pol A1
Synonym(s):
rnase h
About This Item
Recommended Products
biological source
Escherichia coli ( H 560 pol A1)
Quality Level
Assay
100%
form
solution
specific activity
~40000 units/mg protein
packaging
pkg of 100 U
manufacturer/tradename
Roche
technique(s)
cDNA synthesis: suitable
color
colorless
optimum pH
7.5-9.1
solubility
water: miscible
suitability
suitable for molecular biology
NCBI accession no.
application(s)
life science and biopharma
foreign activity
RNase, none detected (up to 10 U with MS- II- RNA)
endonuclease ~10 units, none detected (using lambda-DNA)
nicking activity 10 units, none detected
shipped in
dry ice
storage temp.
−20°C (−15°C to −25°C)
Gene Information
Escherichia coli ... rnhA(946955)
General description
Source: E. coli H560 pol A1
Storage Buffer: 25 mM Tris-HCl, 50 mM KCl, 1 mM dithiothreitol, 0.1 mM EDTA, 50% glycerol (v/v), pH 8.0 (+4°C)
Volume Activity: 1 x 103 U/ml assayed according to Hillenbrand & Staudenbauer.
Application
- In vivo RNA-primed initiation of DNA synthesis
- Elimination of mRNA during second-strand cDNA synthesis
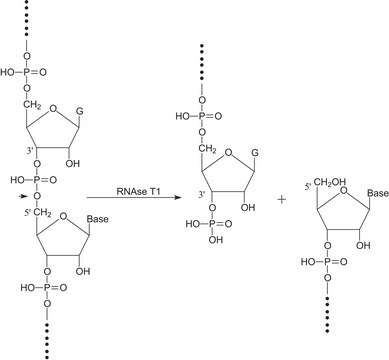
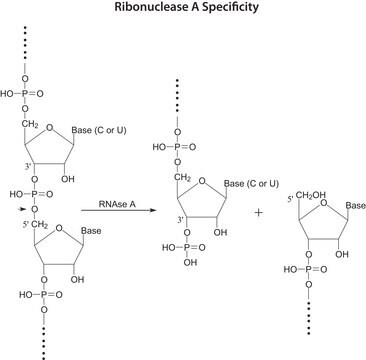
- Site-specific cleavage of RNA
- Detection of RNA:DNA regions in double-stranded DNA of natural origin
- Removal of poly (A) sequences of mRNA if oligo (dT) is present
- RNA extraction and quantitative reverse transcriptase polymerase chain reaction (RT-PCR)
Biochem/physiol Actions
Features and Benefits
- Eliminate potential sources of PCR errors.
- Increase accessibility of primers during subsequent PCR.
Quality
Unit Definition
Volume Activity: Approximately 1 U/μl
Preparation Note
Storage and Stability
Other Notes
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 1
Flash Point(F)
does not flash
Flash Point(C)
does not flash
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service