RPOLT7-RO
Roche
T7 RNA Polymerase
from Escherichia coli BL 21/pAR 1219
Synonym(s):
polymerase
About This Item
Recommended Products
biological source
Escherichia coli (BL 21/pAR 1219)
Quality Level
Assay
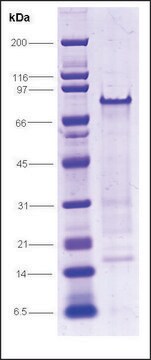
100% (SDS-PAGE)
form
solution
specific activity
≥20 U/μL
mol wt
98 kDa (Single polypeptide chain)
packaging
pkg of 1,000 U (10881767001)
pkg of 5,000 U (10881775001)
manufacturer/tradename
Roche
technique(s)
Northern blotting: suitable
Southern blotting: suitable
hybridization: suitable
color
colorless
pH
7.9 (39 °F)
solubility
water: miscible
suitability
suitable for molecular biology
NCBI accession no.
UniProt accession no.
application(s)
life science and biopharma
foreign activity
Endonucleases 100 units, none detected
Nicking activity 100 units, none detected
RNase 100 units, none detected
storage temp.
−20°C
Gene Information
Escherichia coli ... T7p07(1261050)
Related Categories
General description
Synthesis of hybridization probes: T7 RNA polymerase allows highly efficient production of homogeneously labeled RNA. This labeled RNA may be used as hybridization probes in Southern, northern, and dot blots, as well as in situ hybridizations.
Suitable labels:Transcripts can be nonradioactively labeled with biotin-16-UTP or DIG-11-UTP. They may also be radioactively labeled to high specific activity with [α-32P]- or [α-35S]-labeled nucleotides.
Specificity
T7 RNA polymerase is extremely promoter-specific and only transcribes bacteriophage T7 DNA or DNA cloned downstream of a T7 promoter. Although the T7 and T3 promoter sequences differ only by 3 bp, T7 RNA polymerase only transcribes DNA cloned downstream of its promoter.
Heat inactivation: Stop the reaction by adding 2 μl 0.2 M EDTA (pH 8.0) and/or heat to 65 °C.
Application
- T7 RNA Polymerase can transcribe RNA from cloned DNA templates that are downstream from a T7 promoter. The synthesis can be performed with labeled NTPs to generate highly labeled RNA. Synthesized RNA can be used in many applications, including: RNA or DNA blotting techniques
- In situ hybridization
- RNase protection studies: Transcripts synthesized by the enzyme are used as precursor RNA for studies on RNA splicing and processing.
- Synthesis of capped RNA in vitro with addition of m7GpppG or m7GpppA in excess over GTP or ATP during the transcription reaction. The generated antisense RNA can be introduced into cells to suppress the expression of the corresponding genes.
- Microarray target synthesis
Packaging
Unit Definition
Volume Activity: ≥20 U/μl
Preparation Note
Storage and Stability
Other Notes
40 mM Tris-HCl, pH 8.0 (+20°C), 6 mM MgCl2, 10 mM dithiothreitol, 2 mM spermidine, pH approximately 8.0 (+20°C).
Absence of Endonucleases
1. 1 μg lambda DNA is incubated with T7 RNA polymerase for 4 hours at +37°C in 25 μl test buffer. The number of enzyme units which show no degradation of lambda DNA is >100 U.
2. 1 μg Eco RI/Hind III fragments of lambda DNA is incubated with T7 RNA polymerase for 4 hours at +37°C in 25 μl test buffer. The number of enzyme units which show no alteration of the banding pattern is >100 U.
Absence of Nicking Activity
1 μg pBR322 DNA is incubated with T7 RNA polymerase for 4 hours at +37°C in 25 μl test buffer. The number of enzyme units which show no relaxing of supercoiled structure is >100 U.
Absence of RNases
4 μg MS2 RNA are incubated with T7 RNA polymerase for 4 hours at +37°C in 50 μl test buffer. The number of enzyme units which show no degradation of MS2 RNA is >100 U.
Performance in Transcription Assay
T7 RNA polymerase is function tested in the SP6/T7 Transcription Kit (Cat. No. 10 999 644 001). The incorporation rate in the standard assay with 0.5 μg pSPT19 neo DNA linearized with Eco RI and 50 mCi [alpha-32P] CTP, [400 Ci/mmol (15 TBq/mmol)] gives >50% of the input radioactivity in 20 minutes.
Roche has 10x concentrated RNA Labeling Mixes that are specially designed for DIG- or biotin-labeling. These mixes work well with T7 RNA Polymerase.
Kit Components Only
- T7 RNA Polymerase, in buffer, pH 7.9 ≥20 U/μl
- Transcription Buffer 10x concentrated
Signal Word
Warning
Hazard Statements
Precautionary Statements
Hazard Classifications
Eye Irrit. 2
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 2
Flash Point(F)
does not flash
Flash Point(C)
does not flash
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Articles
T7 RNA Polymerase is a DNA-dependant RNA Polymerase that exhibits a very high specificity for the T7 promoter sequence. The polymerase is useful for synthesizing large amounts of RNA suitable for in vitro translation and anti-sense RNA research.
Understand how mRNA vaccines induce immunity. and read how synthetic mRNA is prepared for vaccine immunogens and other biopharmaceuticals. Find reagents for synthesis of mRNA.
Global Trade Item Number
| SKU | GTIN |
|---|---|
| 10881767001 | 4061837684531 |
| 10881775001 | 4061838687524 |
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service