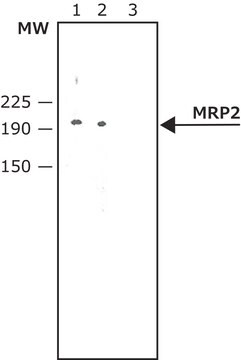
MSP2L
MS PhosphoMix 2 Light
Phosphopeptide Standard for MS
About This Item
Produits recommandés
Qualité
Phosphopeptide Standard for MS
Niveau de qualité
Classe(s) chimique(s) de l'analyte
amino acids, peptides, proteins
Conditionnement
pkg of 200 pmol total phosphopeptides
Technique(s)
HPLC: suitable
LC/MS: suitable
Application(s)
food and beverages
Format
multi-component solution
Température de stockage
−20°C
Description générale
Each of the three phosphopeptides mixes are available in their naturally occurring isotopic abundances (light) or as stable isotope enriched versions (heavy), making the set of products highly amenable to quantitative analyses, allowing users to compare recovery between workflows or techniques.
- Naturally occurring peptide sequences
- Broad range of peptide characteristics
- Complementary product designs
- Available in light and heavy versions
More info and FASTA file
Produit(s) apparenté(s)
Code de la classe de stockage
11 - Combustible Solids
Classe de danger pour l'eau (WGK)
WGK 3
Point d'éclair (°F)
Not applicable
Point d'éclair (°C)
Not applicable
Certificats d'analyse (COA)
Recherchez un Certificats d'analyse (COA) en saisissant le numéro de lot du produit. Les numéros de lot figurent sur l'étiquette du produit après les mots "Lot" ou "Batch".
Déjà en possession de ce produit ?
Retrouvez la documentation relative aux produits que vous avez récemment achetés dans la Bibliothèque de documents.
Notre équipe de scientifiques dispose d'une expérience dans tous les secteurs de la recherche, notamment en sciences de la vie, science des matériaux, synthèse chimique, chromatographie, analyse et dans de nombreux autres domaines..
Contacter notre Service technique