KMM-101NV
KOD One™ PCR Master Mix
Ready-to-use 2X hot-start PCR master mix with a modified KOD DNA polymerase optimized for ultra-fast and convenient high-fidelity PCR
Synonyme(s) :
Fast Hot-start PCR, High-fidelity DNA polymerase, KOD One™ PCR Ready Mix
About This Item
Produits recommandés
Source biologique
Escherichia coli (carrying the cloned UKOD DNA polymerase gene)
Niveau de qualité
Forme
liquid
Utilisation
sufficient for 25 reactions
sufficient for 500 reactions
Caractéristiques
Fast PCR
High Fidelity PCR
dNTPs included
hotstart
Technique(s)
PCR: suitable
Entrée
crude DNA
Conditions d'expédition
dry ice
Température de stockage
−20°C
Description générale
This hot start KOD polymerase is antibody-inactivated with two types of antibodies that prevent the 5′→3′ polymerase activity and 3′→5′ exonuclease activity. Upon heat-activation, the 3′→5′ proof-reading ability of the enzyme generates blunt-end PCR products.
Application
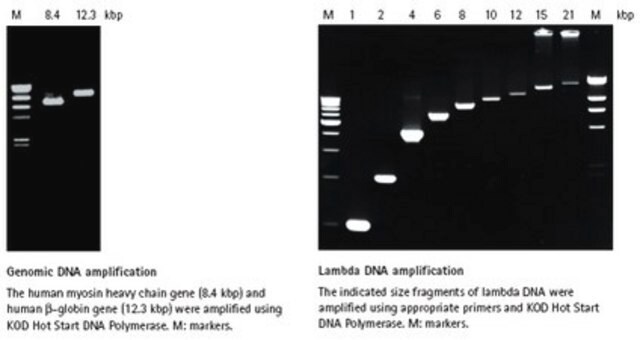
- Genomic DNA amplification
- cDNA amplification
- Direct PCR
- Colony PCR
- Amplification of NGS libraries
- Site direct gene mutation
Compatible Sample Types: Serum, Plasma, Cell, Plant, soil extraction, DNA, nail, hair etc.
Sample Volume Needed: about 50ng DNA
Caractéristiques et avantages
- Fast: Amplifies the targets using the following very short conditions:
o 1~ 10 kb: 5 sec/ kb
o 10 kb~: 10 sec/ kb
- Ready-to-use: 2X premixed solution of UKOD DNA polymerase, buffer, dNTPs, and MgCl2.
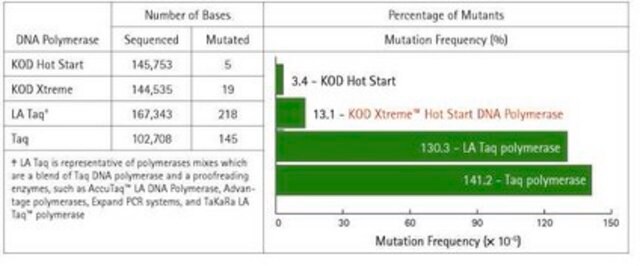
- High Fidelity: Approximately 80-fold higher fidelity than conventional Taq DNA polymerase
- High Efficiency: Enables high throughput PCR and ultra-fast PCR cycling conditions with an extension time of as quick as 5 sec/kb.
- Crude Sample PCR: Effective for direct PCR amplification of crude samples such as biological samples, food samples, soil extract, etc.
- High flexibility: Amplifies templates containing uracils (dU). Also, amplifies fragments using degenerate primers with inosines (dI) and uracils (dU)
Conditionnement
KMM-101NVS - 0.25ML is sufficient for 25 reactions at 20 μL each or for 10 reactions at 50 μL each.
Autres remarques
Informations légales
Mention d'avertissement
Warning
Mentions de danger
Conseils de prudence
Classification des risques
Met. Corr. 1
Code de la classe de stockage
8B - Non-combustible corrosive hazardous materials
Classe de danger pour l'eau (WGK)
WGK 2
Point d'éclair (°F)
Not applicable
Point d'éclair (°C)
Not applicable
Certificats d'analyse (COA)
Recherchez un Certificats d'analyse (COA) en saisissant le numéro de lot du produit. Les numéros de lot figurent sur l'étiquette du produit après les mots "Lot" ou "Batch".
Déjà en possession de ce produit ?
Retrouvez la documentation relative aux produits que vous avez récemment achetés dans la Bibliothèque de documents.
Contenu apparenté
KOD One™ PCR Master Mix overview for ultra-fast PCR with high specificity, fidelity, and yield
Notre équipe de scientifiques dispose d'une expérience dans tous les secteurs de la recherche, notamment en sciences de la vie, science des matériaux, synthèse chimique, chromatographie, analyse et dans de nombreux autres domaines..
Contacter notre Service technique