SCC117
Histone H2B-GFP expressing HeLa Cell Line
Human
Synonym(s):
Histone H2B
About This Item
Recommended Products
product name
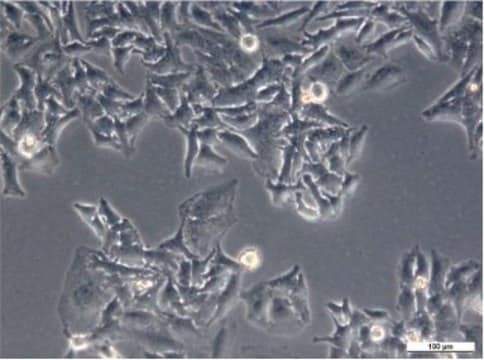
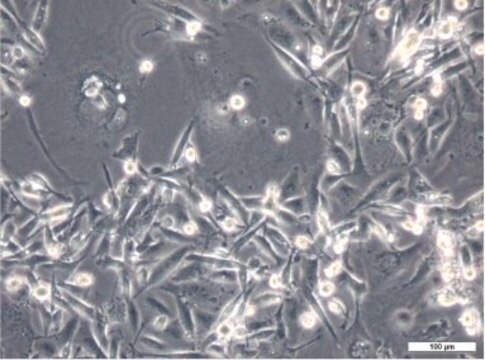
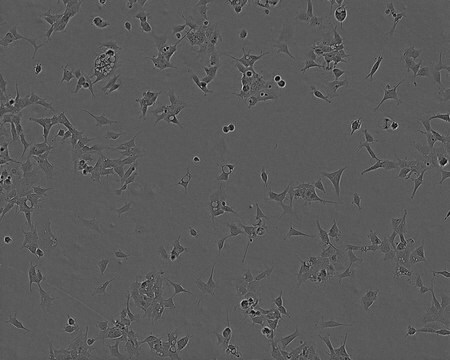
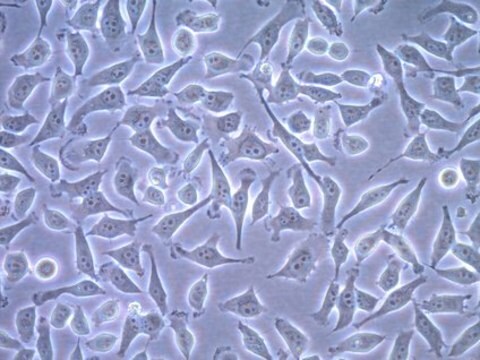
Histone H2B-GFP expressing HeLa Cell Line, The Histone H2B-GFP expressing HeLa cell line is a useful cell model for imaging and analysis of chromosome dynamics.
biological source
human
Quality Level
technique(s)
cell based assay: suitable
cell culture | mammalian: suitable
shipped in
ambient
General description
Cell Line Description
Application
Apoptosis & Cancer
Epigenetics & Nuclear Function
Cell Cycle, DNA Replication & Repair
Chromatin Biology
Histones
Quality
• Cells are tested by PCR and are negative for HPV-16, HPV-18, Hepatitis A, B, C and HIV-1 & 2 viruses.
• Cells are negative for mycoplasma contamination.
• Each lot of cells are genotyped by STR analysis to verify the unique identity of the cell line.
Storage and Stability
Disclaimer
Signal Word
Danger
Hazard Statements
Precautionary Statements
Hazard Classifications
Acute Tox. 3 Oral
Storage Class Code
6.1C - Combustible acute toxic Cat.3 / toxic compounds or compounds which causing chronic effects
WGK
WGK 2
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service