Salt Selection and Buffer Preparation
Salts
In HIC the binding process is more selective than the elution process, so it is essential to optimize the conditions of the start buffer. The correct choice of salt and salt concentration are the most important parameters that influence capacity and final selectivity. The objective is to optimize conditions to achieve the required selectivity to bind the target protein(s) and ensure that the majority of impurities pass through the column.
The influence of different salts on hydrophobic interaction is explained in Chapter 1. In practice sodium, potassium or ammonium sulfates effectively promote ligand-protein interactions in HIC and have a stabilizing influence on protein structure. Hence the most commonly used salts are (NH4)2SO4, Na2SO4, NaCl, KCl and CH3COONH4. Figure 1 shows an example of how different salts can affect selectivity. Here the best resolution of four standard proteins was obtained using 1.7 M ammonium sulfate in the start buffer.
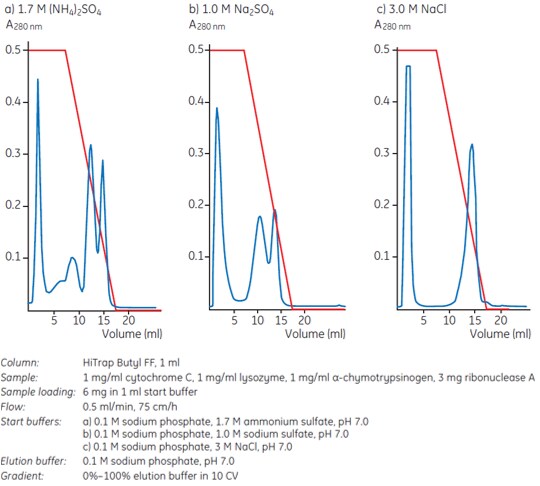
Figure 1. Different salts influence selectivity: elution takes place in order of increasing elution volumes: cytochrome C, lysozyme, ribonuclease A, α-chymotrypsinogen.
As with media selection, the choice of salt for a HIC separation can be a matter of trial and error since each salt differs in its ability to promote hydrophobic interactions. As the concentration of a salt increases, the amount of protein bound will increase almost linearly up to a specific salt concentration and continue to increase in an exponential manner at higher concentrations.
- At a given concentration, ammonium sulfate often gives the best resolution when compared to other salts and can be used at concentrations up to 2 M.
- Concentrations up to 3 M are usually required when using sodium chloride.
- Sodium sulfate is a very good salting-out agent, but problems with protein solubility may exclude its use at high concentrations.
- Ammonium sulfate is not recommended for working at pH values above 8.0.
Salt concentration
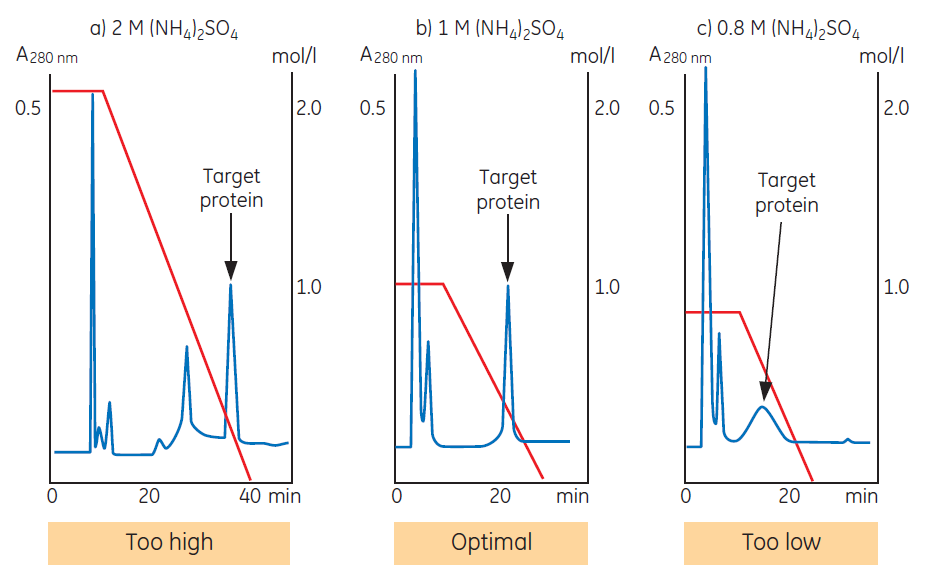
Figure 2. Salt concentration in the start buffer influences selectivity and resolution.
Figure 2 shows the influence of salt concentration on selectivity and resolution. In this example the target protein is the last peak to elute. The selectivity of the medium is satisfactory since the protein elutes within the gradient and is well resolved from contaminants. In (a), the target protein elutes in a sharp zone, but late in the gradient. Lowering the initial salt concentration (b) gives similar resolution, but ensures that contaminants that bound during earlier runs (when a higher salt concentration was used) now elute during the initial wash step. Only the target protein is bound, reducing the risk of a contaminant co-eluting with the target protein and increasing the capacity of the column for the target protein. A run performed at even lower initial salt concentration (c) shows good selectivity but poor efficiency for the target protein. The sample is not bound strongly enough during sample application, resulting in significant peak broadening during elution.
- If the target molecule elutes too late or not at all and switching to a different medium is not possible, try binding in 50% less salt.
- Some proteins begin to precipitate at high salt levels. The salt concentration in the start buffer may need to be reduced to prevent precipitation during the run. Loading the sample repetitively in small amounts can also help to avoid losing yield due to precipitation.
Buffer Ions and pH
Selection of buffering ions is not critical for hydrophobic interaction. Phosphate buffers are most commonly used.
The pH chosen must be compatible with protein stability and activity, and it is advisable to check for optimum pH conditions for each specific application. However, between pH 5–8.5, pH values have very little significance on the final selectivity and resolution of a HIC separation. An increase in pH weakens hydrophobic interactions and retention of proteins changes more drastically at pH values above 8.5 or below 5.0.
- Check for stability at the pH and salt concentrations used during the separation, especially if recovery of biological activity is a priority. Avoid extreme changes in pH or other conditions that may cause inactivation or even precipitation.
- Use a buffer concentration, typically 20–50 mM, that is sufficient to maintain buffering capacity and pH during sample application and changes in salt concentration.
- Transfer the purified protein into a volatile buffer if the product is to be lyophilized. Table 3 lists suitable volatile buffer systems.
- Prepare buffers at the same temperature at which they will be used to ensure the correct pH.
- Filter buffers and samples after all salts and additives have been included. Use high-quality water and chemicals. Use 1 μm filters for media with particle sizes above 90 μm, 0.45 μm filters for 34 μm particles, and 0.22 μm filters for particles below 15 μm or when sterile or extra-clean samples are required. To avoid formation of air bubbles in a packed column and to ensure reproducible results, the column and buffers should be at the same temperature when preparing for a run.
- For samples with unknown hydrophobic properties, try the following:
Start buffer: 1.5 M ammonium sulfate, 50 mM sodium phosphate, pH 7.0
Elution buffer: 50 mM sodium phosphate, pH 7.0
Buffer additives
Additives can be used to improve selectivity and resolution, for example when a protein binds too strongly to a HIC medium. However, if used at high concentrations, there is a risk of inactivating and/or denaturing the target protein. Additives can influence a separation by improving protein solubility, modifying protein conformation and promoting elution of bound proteins. Water-miscible alcohols, detergents, and chaotropic salts (limited use) are the most widely used additives in HIC separations. Typical additives are shown in Table 4.
Run blank elution gradients with additives included in order to check their effect on the elution profile (that is, perform a run but do not load any sample).
Para continuar lendo, faça login ou crie uma conta.
Ainda não tem uma conta?