Detergent Solubilization of Membrane Proteins
How Do Detergents Solubilize Membrane Proteins?
Detergents are widely used in biochemistry, cell biology and molecular biology. Common applications include cell lysis, solubilization of membrane proteins and lipids, protein crystallization, and reduction of background staining in blotting experiments.
Even though studying membrane proteins is a major challenge in protein biochemistry, they remain an important area of study due to their significant biological and pharmacological relevance. Understanding the structure and function of membrane proteins requires their careful isolation in the native form in a highly purified state. Since micelle-forming detergents provide an amphipathic environment that can mimic lipid bilayers, their use as solubilizing agents is essential for functional and structural investigations.
Biological membranes consist of phospholipids that contain two hydrophobic groups connected to a polar head. This molecular architecture allows phospholipids to assemble as lipid bilayers in which the hydrophobic chains face each other while the polar head groups face outward to the aqueous milieu (Figure 1). Proteins and lipids are embedded in this bilayer forming the fluid mosaic model (Figure 2) which was first proposed by Singer and Nicolson in 1972.
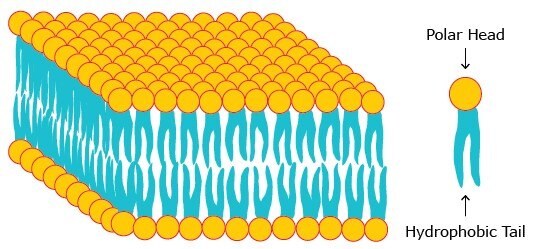
Figure 1. A phospholipid bilayer
Proteins are held in the lipid bilayer by hydrophobic interactions between the lipid tails and hydrophobic protein domains.1 These integral membrane proteins (IMPs) (Figure 2) are not soluble in aqueous solutions as they aggregate to protect their hydrophobic domains, but are soluble in detergent solutions as micelles formed by detergents are analogous to the bilayers of the biological membranes.2 Proteins are incorporated into these micelles via hydrophobic interactions. Hydrophobic regions of membrane proteins, normally embedded in the membrane lipid bilayer, are now surrounded by a layer of detergent molecules and the hydrophilic regions are exposed to the aqueous medium. This keeps the membrane proteins in solution. Complete removal of detergent could result in aggregation due to the clustering of hydrophobic regions and, hence, may cause precipitation of membrane proteins.3
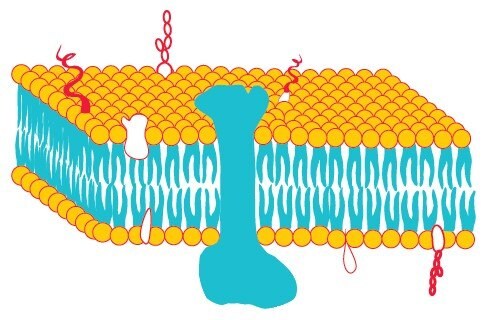
Figure 2.Fluid-mosaic model of a biological membrane
Membrane solubilization by detergents can be described as a three stage process where the detergent-lipid-protein ratio is an important factor (Figure 3).4
When low concentrations of a detergent are added to biological membranes (a), the detergent monomers (shown in red with single tails) perturb the membrane structurally by partitioning into the lipid bilayer (b). At concentrations equal to, or higher than the detergent’s CMC, the lipid bilayer becomes saturated with detergent molecules and breaks apart generating lipid-protein-detergent mixed micelles (c). 8 A detergent/protein ratio of around 1-2 (w/w) is believed to be sufficient to solubilize IMPs to form lipid-protein-detergent mixed micelles.5 A further increase of detergent concentration causes progressive delipidation of the lipid-protein-detergent mixed micelles. This leads to the formation of lipid/detergent and protein/detergent mixed micelles (d). 8 A solubilized IMP in a complex with a bound detergent is called a protein-detergent complex, PDC.6 Typically, a detergent/protein ratio of around 10 (w/w) or higher will lead to complete delipidation.7
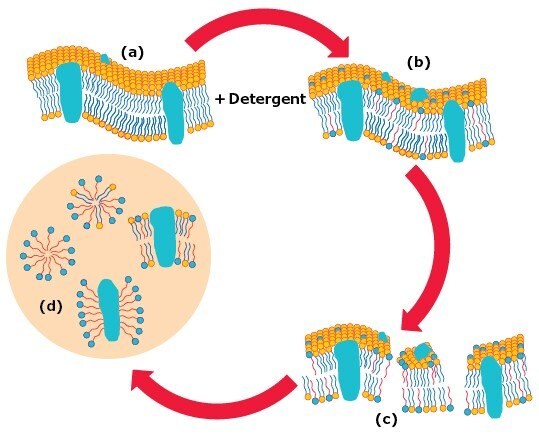
Figure 3.Stages in the solubilization of biological membranes by detergents.
Removal of Unbound Detergents
Excess detergent is normally employed in solubilization of membrane proteins to ensure complete dissolution of the membrane and provide a large number of single protein molecule containing micelles. However, for further physicochemical and biochemical characterization of membrane proteins, it is often necessary to remove the unbound detergent.
The following is a brief description of four commonly used methods for detergent removal that take advantage of the general properties of detergents: e.g., hydrophobicity, CMC, aggregation number, and the charge.
Hydrophobic Adsorption
This method exploits the ability of detergents to bind to hydrophobic resins. Generally, a detergent-containing solution is mixed with a specific amount of the resin, and the mixture is allowed to stand at 4° C or room temperature. The resin with the bound detergent can be removed by centrifugation or filtration. This technique is effective for the removal of most detergents and especially suitable for detergents with low CMCs.8 If the adsorption of the protein to the resin is of concern, the resin can be included in a dialysis buffer and the protein dialyzed.
Gel Chromatography
Gel chromatography takes advantage of the difference in size between protein-detergent, detergent-lipid, and homogeneous detergent micelles. In most situations, protein-detergent micelles elute in the void volume.
The elution buffer should contain a detergent below its CMC value to prevent protein aggregation and precipitation. Since this method is based on separation based on size, parameters that influence micellar size (ionic strength, pH, and temperature) should be kept constant from experiment to experiment to obtain reproducible results.2
Dialysis
When detergent solutions are diluted below the CMC, the micelles are dispersed into monomers. Monomers are usually significantly smaller than micelles, and can be easily removed by dialysis. Dialysis is the most common form of detergent removal and typically requires dialyzing the protein detergent mixtures against detergent-free buffer (in about 200-fold excess). If a large dilution is not practical, micelles can be dispersed by other techniques such as the addition of bile acid salts. Dialysis is more practical with detergents having high CMCs and works best for those with low molecular weight/small cross-sectional area.2
Ion-Exchange Chromatography
This method exploits the differences in charge between protein-detergent micelles and protein-free detergent micelles. When non-ionic or zwitterionic detergents are used, conditions can be chosen to retain protein-containing micelles on the ion-exchange resin, letting protein-free micelles pass through. The adsorbed protein can then be eluted by changing the ionic strength or pH of detergentfree buffer, or by washing with an ionic detergent.2
References
Pour continuer à lire, veuillez vous connecter à votre compte ou en créer un.
Vous n'avez pas de compte ?