Chromatofocusing: Principles and Methods
Use chromatofocusing as a polishing step for partially purified samples. The fewer components in the sample, the better the chance for a well-resolved separation of individual proteins. For specific applications, chromatofocusing can resolve molecules where pI values differ by as little as 0.02 pH units. This high resolving ability can be particularly useful for the separation of very similar substances.
Use chromatofocusing for high resolution, analytical separations.
Use chromatofocusing as a complementary technique when proteins have not been resolved by other chromatography techniques such as charge (using ion exchange), size or hydrophobicity.
Media selection
- Use prepacked, Mono P for fast separations (up to ten times faster than PBE).
- Use Mono P 5/200 GL for highest resolution separations.
- Use Mono P 5/50 GL rapid scouting of elution conditions (pH gradients) or for separations not requiring highest resolution.
- Use PBE 94 for scaling up from Mono P. Note that, under the same separation conditions, resolution may be lower due to the longer diffusion times within larger particles. Reoptimization of the gradient and flow rate should give similar resolution.
- Use PBE 118 when pH gradients above pH 9 are required.
Mono P is based on the same matrix as Mono Q and Mono S. The 10 μm MonoBeads particles are substituted with tertiary and quaternary amines and the small bead size contributes significantly to the high resolution that can be achieved. Polybuffer exchangers, PBE 118 and PBE 94, are based on Sepharose CL-6B, a cross-linked agarose matrix. The charged secondary, tertiary and quaternary amine groups are coupled to monosaccharide units in the 90 μm Sepharose CL-6B particles by ether linkages. For all chromatofocusing media the charged groups have been selected to give an even buffering capacity across a broad pH range. The buffering capacity and titration curves for PBE 118, PBE 94 and Mono P are shown in Figures 79 and 80.

Figure 79.Titration of 10 mL aliquots of PBE 118 and PBE 94 in 1 M KCl against NaOH shows the smooth buffering capacity of these media over a broad pH range.
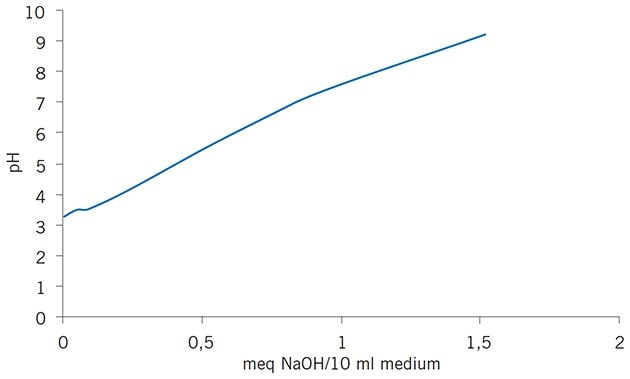
Figure 80.Titration curve of Mono P column shows smooth buffering capacity.
Buffer selection
- Use Polybuffer 74 for any pH gradient between 7 and 4. Use a pH gradient 7–4 when starting to work with proteins of unknown pI.
- Use Polybuffer 96 for pH gradients that should begin above pH 7 e.g. when the proteins of interest have a pI above or close to 7.
- Use Pharmalyte pH 8–10.5 when higher pH values are required.
- Refer to section ‘Selection of pH gradient and buffers’ for further details.
Polybuffers are specifically designed to form pH gradients by titration of the charged groups on PBE 118, PBE 94 and Mono P media. Polybuffers are mixtures of selected amphoteric buffering substances of different pI and pKa values. Each mixture is designed to give an even buffering capacity across a wide pH range, in order to generate the required linear pH gradient. The titration curves for Polybuffer 96 and Polybuffer 74 are shown in Figure 81.

Figure 81.Titration of 2 mL Polybuffer with 0.1 M NaOH.
Purification options |
|---|
*Appendix 5 convert linear flow (cm/hour) to volumetric flow rates (mL/min) and vice versa.
**Working pH range refers to the pH interval where the medium binds protein as intended or as needed for elution without adverse long term effects.
***Maximum operating back pressure refers to the pressure above which the medium begins to compress.
Purification examples
High resolution
The potential for high resolution separation of proteins with otherwise similar properties makes chromatofocusing an excellent technique for the study of different isoforms. For example, haemoglobins can be separated into 4 distinguishable sub-groups, each with a different isoelectric point (Figure 82). The isoelectric points of sub-groups A and F differ by only 0.05 pH units, yet the peaks are well resolved. The focusing effect of the linear pH gradient and excellent peak symmetry (achieved by using small, uniform particles in a well-packed column) combine to give a high resolution separation.

Figure 82.Chromatofocusing of haemoglobin variants.
Under ideal conditions with a very well packed column, Polybuffer exchangers can give almost as good resolution as Mono P. Figure 83 shows an example in which model proteins that differ by as little as 0.02 pH units in their pI values have been resolved on a Polybuffer exchanger.

Figure 83.Chromatofocusing of standard protein mixture.
High selectivity
Figure 84 demonstrates the high selectivity of chromatofocusing that enables detection of subtle changes in individual proteins. In this application researchers were able to follow the desialylation of transferrin as sialic acid groups were removed by neurominidase.

Figure 84.Following the desialylation of transferrin.
Polishing step
Figure 85 shows an example of chromatofocusing being used for polishing in a purification strategy (in a polishing step most impurities have been removed and the objective is to achieve final purity by removing any trace impurities or closely related substances). In this example highly pure leukotriene A4 hydrolase was required for functional and structural studies. The enzyme was expressed as (His)6-tagged LTA4 hydrolase in E. coli. After harvesting the cells and removing nucleic acids, nickel ion affinity chromatography was used as a specific protein capture step and the eluted fraction subjected to a polishing step on Mono P.
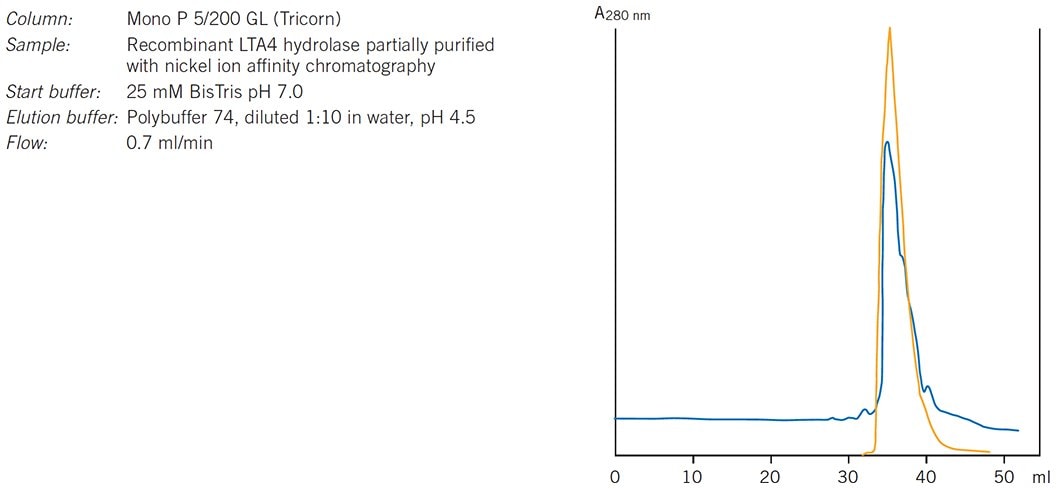
Figure 85.Purification of leukotriene A4 hydrolase. Results courtesy of Eva Ohlson, Karolinska Institutet, Stockholm, Sweden.
Packing a column
Prepacked columns are likely to give the highest resolution and the most reproducible results, particularly with the MonoBead matrix used in Mono P 5/50 GL and Mono P 5/200 GL. Efficient packing of a chromatofocusing column is critical to achieve the best results. Follow carefully the instructions for column packing given in Appendix 3 when packing PBE 94 or PBE 118.
Pack columns in the start buffer to be used for the separation (see Table 10, 11, 12 or 13) using long, narrow columns such as Tricorn 10/300. The amount of medium required will depend on the amount of sample, the nature of the sample and contaminants and the degree of resolution required. For most separations 20–30 mL Polybuffer exchanger will be sufficient to separate from 1–200 mg protein/pH unit in the gradient.
Degas the start buffer and the slurry in order to avoid air bubbles which can interfere with the separation.
Check column efficiency as described in Appendix 3. It may be worth checking the packing using a colored marker. Use bovine cytochrome c which is strongly repelled from the medium due to its high isoelectric point (pI=10.5). The progress of the protein band through the column can be visualized in order to check for any distortion caused by poor column packing or air bubbles.
Selection of pH gradient and buffers
No gradient forming equipment is required for chromatofocusing since the gradient is generated on the column as buffer and medium interact. The upper limit of the gradient is defined by the pH of the start buffer and the lower limit of the gradient is defined by the pH of the elution buffer. Polybuffers perform best over pH intervals of 3 pH units or less and the narrowest pH intervals are likely to give the highest resolution.
The gradient volume is defined by the strength of the elution buffer (low buffer concentrations give gradual pH changes and good separations between peaks). The optimal gradient volume may need to be determined by experimentation. Recommended gradient volumes and buffers are indicated in Tables 10, 11, 12 and 13. The pre-gradient volume is the volume of elution buffer that passes through the column before the pH begins to decrease, hence the total volume of buffer required is always greater than the gradient volume.
Start buffers: avoid the use of polyvalent buffers. Use monovalent or amphoteric buffers with a net charge > 0 and < +1and ionic strength of 25 mM. Set the pH of the start buffer to 0.4 pH units above the desired pH in order to compensate for fluctuations in pH at the start of the run. These fluctuations are caused by slight differences in the conductivity of start and elution buffers.
Avoid buffers at concentrations below 25 mM as they will require larger gradient volumes with longer elution times and give broader, less concentrated protein peaks. More concentrated buffers would give steeper gradients, but could result in loss of resolution as peaks would be eluted closer together.
Gradients ending at pH 9 are not recommended when using Mono P since this medium has low buffering capacity above pH 9.0.
Gradients for proteins of known pI
Choose a pH interval so that the protein of interest is eluted after running 30–50% of the pH gradient. Narrow pH intervals will give optimal resolution.
Gradients for proteins of unknown pI
Since most proteins have isoelectric points in the range of 7–4, start with pH interval 7–4 on PBE 94 or Mono P. Proteins with a pI above 7 will pass through the column and can be collected and re-run at a higher pH if desired. Once a suitable pH range is established, select the most suitable pH gradient and compatible medium to achieve the optimal resolution.
Save time by using the shorter Mono P 5/50 GL column to test for a suitable pH gradient before transferring to Mono P 5/200 GL to achieve the highest resolution. Routine separations on Mono P 5/50 GL may also be satisfactory if highest resolution is not essential.
*Use a saturated solution of iminodiacetic acid to adjust the pH of the buffer.
*Use a saturated solution of iminodiacetic acid to adjust the pH of the buffer.
Selection of counter-ions
As can be seen in the buffer tables, the most commonly used counter-ion is chloride. Other monovalent counter-ions can be used, but it is essential that these anions have a pKa at least two pH units below the lowest point of the chosen gradient.
Acetate is not recommended as a counter-ion for Polybuffer 74 because it has a high pKa. Multivalent counter-ions with a net charge below -1 are not recommended. Note that iminodiacetic acid is a multivalent ion.
Bicarbonate ions can result from the presence of atmospheric CO2 or badly stored buffers. All high pH, amine-containing buffers adsorb atmospheric CO2 and so generate bicarbonate ions which can disrupt a pH gradient by causing a fluctuation or plateau in the region of pH 5.5–6.5, depending on conditions.
These effects are most marked with Polybuffer 96 in pH gradients ending at pH 6, and can be minimized by using acetate as the counter-ion or setting the lower limit to pH 6.5. Adsorption of CO2 can be minimized by storing solutions under nitrogen in tightly sealed bottles and at 3–8 ° C in the dark.
Buffer preparation
Use high quality water and chemicals. Filter buffers through 0.45 μm or 0.22 μm filters under vacuum to ensure that the solutions are thoroughly degassed. The presence of air bubbles in the column can significantly interfere with resolution.
Ensure that all buffer components are stored correctly to avoid adsorption of CO2 (especially those containing amines). Use fresh Polybuffer from a previously unopened bottle whenever possible. Store previously opened solutions under nitrogen in tightly sealed bottles at +4 °C and in the dark to minimize adsorption of CO2.
Prepare and use all buffers carefully at the same temperature to ensure correct pH and ionic strength. Use fresh buffers whenever possible. Buffers stored under refrigeration must reach the temperature at which they were prepared before running a separation.
The concentration of the start buffer is especially important when working at low ionic strength where microenvironments can vary by as much as 1 pH unit. Alterations in ionic strength may occur if readjustments to pH are made when buffers have been overtitrated with acid. Ensure that start and elution buffers are at the same low ionic strength in order to avoid large pH changes at the beginning or end of a pH gradient.
- Select start and elution buffers from Tables 10, 11, 12 or 13 according to the pH gradient required and the medium to be used.
- Calculate the required buffer volumes according to the column volume used. For the elution buffer, dilute Polybuffer and/or Pharmalyte in distilled water to approximately 95% of the final volume required.
- Titrate buffers to the correct pH with the listed acid (e.g. 1–2 M or saturated iminodiacetic acid). Always perform the titration on a maximum volume before adding a few milliliters of water to reach the final volume.
- When the final pH is reached, add distilled water to bring the total volume to 100 mL.
For shallower gradients within the same pH interval, prepare the elution buffer as normal, but dilute to a larger volume. Note that proteins elute with increased volumes when using diluted eluents so pre-gradient and total volumes also need to be increased.
Sample preparation
Correct sample preparation is extremely important. Simple steps to clarify a sample before applying it to a column will avoid the risk of blockage, reduce the need for stringent washing procedures and extend the life of the packed medium. Appendix 1 contains an overview of sample preparation techniques.
Samples must be clear and free from particulate matter, particularly when working with Mono P. For small sample volumes a syringe-tip filter of cellulose acetate or PVDF can be sufficient for sample filtration.
If the pH of the sample is too low or different buffers are used, both gradient and resolution may be affected. Depending on the sample volume, use a HiTrap Desalting or HiPrep 26/10 Desalting column to remove high salt concentrations and/or transfer the sample into start buffer. The pH of the sample is not critical if the buffer concentration of the sample is very low. It may be possible to simply dilute the sample if pH and final volume are not critical.
Since chromatofocusing is a binding technique, the volume of the sample is not significant as long as all of the sample can be applied and focused before the proteins of interest begin to elute. As a general rule, do not load a sample volume that is greater than half of the total column volume.
Performing a separation
Recommended flow: 0.5–1.5 mL/min, 305–450 cm/h (Mono P 5/50 GL, Mono P 5/200 GL), 30–40 cm/h (PBE 118, PBE 94).
Start and elution buffers: see Tables 10, 11, 12 or 13.
Monitor eluent at 280 nm since Polybuffers absorb at wavelengths below 280 nm. Monitor pH throughout the run as fluctuations can occur that may affect the separation, for example, due to the presence of CO2 in the buffer.
Using a column for the first time or after long term storage:
- Inject 0.5 column volumes 5 M NaOH
- Equilibrate with start buffer until the buffer leaving the column is at the same pH as the start buffer.
- Apply 0.5 column volumes 2 M salt solution containing the same anion as used in the acid for pH titration of the start buffer.
- Re-equilibrate the column with start buffer until eluent leaving the column is at the start pH.
To check for pH fluctuations or other disturbances caused by incorrect buffer conditions or contaminants, consider performing a blank run using elution buffer (total volume as indicated in Tables 10, 11, 12 or 13).
Using a column for repeated runs:
- Equilibrate with start buffer until buffer leaving the column is at the same pH as the start buffer. See Table 10, 11, 12 or 13 for recommended pre-gradient volumes.
- Adjust sample to pH of start buffer or exchange sample into start buffer using, for example, a HiTrap Desalting column.
- Apply elution buffer. Note that there is a pre-gradient volume that will be eluted before the pH gradient begins (see Tables 10, 11, 12 or 13 for typical pre-gradient volumes).
- Apply sample.
- Apply elution buffer using gradient volumes as indicated in Tables 10, 11, 12 or 13. Check for pH fluctuations or other disturbances during elution.
- Wash with 2 column volumes 2 M NaCl to elute any material still bound to the column.
- Re-equilibrate with 5 column volumes start buffer until UV absorbance and pH/conductivity values are stable.
To maintain a clean column, inject 0.5 column volumes of 1 M NaOH every tenth run (or more frequently if required). The injection of NaOH can be followed by 0.5 column volumes of 75% acetic acid.
If the fractions are to be analyzed by reversed phase chromatography, Polybuffer may interact with the pairing ions used during the run. If the pairing ions are non-hydrophobic Polybuffer will elute in the void volume and the retained samples can be eluted with a gradient of organic solvent. However, if the pairing ions are very hydrophobic, Polybuffer will be retained on the column and give an absorption peak at wavelengths below 280 nm during an elution.
If organic solvents must be used, note the following:
- Before running a column, check the solubility of sample and all buffers.
- The pH interval required may be altered since the pKa values of the charged groups in the buffers, Polybuffer and chromatofocusing medium will be increased.
- Perform a blank gradient to ensure maintenance of a linear pH gradient. Linearity is likely to vary more at the high and low end of a pH interval 9–4. Adjust to the highestpH in the useful buffering range of the start buffer substance and adjust the elutionbuffer to approximately 0.5 pH units higher than recommended in the buffer Tables 10,11, 12 or 13.
Optimization
- If results suggest that there may be a problem of sample solubility during the separation (see Troubleshooting) include additives such as betaine (10% w/v), taurine (4% w/v) or glycerol (1–2%) in the start and elution buffers to improve solubility. These additives should not affect the separation, but may need to be removed at a later stage thus adding an extra step to the purification strategy.
- To improve resolution:
–dilute Polybuffer (up to 1:20), but note that this will increase the pre-gradient and total gradient volumes so the time at which components elute will change and the volume of an eluted peak may increase
–decrease sample load
–decrease flow (flow rates as low as 0.25 mL/min have been used successfully on Mono P columns) - To improve selectivity, use a shallower gradient (increase gradient volume), but note that this will result in broader peaks because each pH interval will occupy a larger space on the column. However, the resolution within a pH interval will increase.
- To change selectivity, use alternative buffers, salts or counter-ions.
If resolution is satisfactory, it may be possible to increase flow rate in order to speed up the separation. Flow rates up to 1.5 mL/min have been used successfully with Mono P columns without loss of resolution. Figure 86 shows the effect of increasing flow on the separation of a test mixture of proteins on PBE 118.

Figure 86.Effect of flow on resolution during chromatofocusing.
Troubleshooting
A common difficulty encountered during chromatofocusing is that some proteins precipitate when they reach high concentrations at or near their isoelectric point. On-column precipitation causes symptoms such as increased back pressure, apparent loss of sample and even blockage of the column.
To avoid precipitation it may be possible to reduce the sample load so that the proteins do not reach sufficiently high concentrations, include additives such as betaine to improve sample solubility or to alter the pH range of the separation to avoid precipitation. Alternatively, remove the proteins causing the problem by using another chromatography technique before chromatofocusing.
If the protein of interest precipitates reversibly, then it will elute later than expected. If the protein of interest precipitates irreversibly at its isoelectric point, then chromatofocusing is not a suitable technique for purification.
A second factor that can easily affect results is the linearity of the pH gradient. The presence of CO2, especially in the start buffer, can distort the pH gradient and disrupt the separation. It is essential to monitor the pH gradient throughout the run to ensure a linear pH gradient and a satisfactory separation. Figure 87 demonstrates the significant effect that excess CO2 can have on the formation of a pH gradient. Store buffers under nitrogen or argon (to minimize the adsorption of CO2 which occurs with high pH solutions) and always degas before use.

Figure 87.A plateau in a pH gradient during chromatofocusing caused by excess CO2 in the start buffer
These difficulties and others that may occasionally be encountered during chromatofocusing are listed in the troubleshooting table that follows.
*Polar organic solvents such as methanol, ethanol and acetonitrile can be used at concentrations from 0–20%, but this may cause some proteins to lose their biological activity. The presence of organic solvents may also affect the low end of a linear pH gradient.
Removing Polybuffer
For most practical applications it is not necessary to remove Polybuffer since the amount that elutes with any sample is extremely low. Polybuffers do not interfere with enzyme assays or amino acid analysis, but they may interfere with certain protein assays such as Lowry.
Polybuffers can be removed from protein samples using a gel filtration medium with a suitable fractionation range. Figure 88 shows separation of Polybuffer and protein using either Sephacryl™ S-100 HR or Superdex 75. In this example Sephacryl S-100 gives the best separation between the protein myoglobin and Polybuffer. It is recommended to follow any separation by monitoring the absorbance of Polybuffer (A215 nm) as well as the absorbance of the eluting protein (A280 nm, A254 nm or A215 nm) in order to optimize the running conditions and ensure effective separation. For practical and theoretical information on gel filtration chromatography, refer to the handbook Gel Filtration–Principles and Methods available from Cytiva.

Figure 88.Separation of Polybuffer from protein using gel filtration.
Cleaning
Mono P 5/50 GL, Mono P 5/200 GL, PBE 94, PBE 118
Since certain proteins may precipitate at or near their isoelectric point, blockage of the top filter on a chromatofocusing column is often the most common reason for an increase in back pressure. Reverse the flow direction and run through 2 column volumes of elution buffer at 0.5 mL/min (Mono P) or 30 cm/h (PBE). Return to normal flow direction, run through 5 column volumes of elution buffer.
To remove severe contamination (often indicated by an increase in column back pressure) proceed as follows:
Reverse flow direction and run the following sequence of solutions at a flow rate of 0.25–0.50 mL/min.
- Wash with 4 column volumes 1 M NaCl.
- Rinse with 2 column volumes distilled water.
- Wash with 4 column volumes 1 M NaOH.
- Rinse with 2 column volumes distilled water.
- Wash alternately with 0.5 column volumes 0.1 M HCl (PBE) or 1 M HCl (Mono P) and 2 column volumes distilled water until the elution profile is constant.
- Wash with 4 column volumes 1 M NaCl.
- Reverse flow direction and re-equilibrate column in start buffer.
If back pressure remains high, change top filter.
Save time by monitoring any cleaning procedure to check for elution of contaminants.
Depending on the nature of the contaminants, the following cleaning solutions can also be used: 100% isopropanol, 20% acetonitrile, 2 M NaOH, 75% acetic acid, 20% ethanol, 100% methanol or up to 6 M guanidine hydrochloride, cationic or non-ionic detergents. Always rinse with at least 2 column volumes of distilled water after using any of these cleaning solutions. When using organic solvents, wash the column using sawtooth gradients e.g. run from 0–100% solvent in 5 column volumes then from 100–0% in 5 column volumes, including 1% trifluoroacetic acid in the water and organic solvent.
If column performance is still not restored, inject a solution of 1 mg/mL pepsin in 0.1 M acetic acid containing 0.5 M NaCl and leave overnight at room temperature or one hour at 37 ºC. Depending on the contaminant, other enzymes may also be used, e.g. DNase. After any enzymatic treatment, repeat the steps to remove severe contamination described previously.
Media characteristics |
|---|
*Long term pH stability refers to the pH interval where the medium is stable over a long period of time without adverse side effects on the chromatography performance.
Short term pH stability refers to the pH interval for regeneration, cleaning-in-place and sanitization procedures. All ranges are estimates based on the experience and knowledge within Cytiva.
Chemical stability
Mono P 5/50 GL, Mono P 5/200 GL
Mono P is stable in all commonly used, aqueous buffers in the range of pH 2–12, and in the presence of additives such as denaturing agents (8 M urea or 6 M guanidine hydrochloride) and non-ionic or cationic detergents.
Avoid oxidizing agents and anionic detergents.
PBE 94, PBE 118
Polybuffer exchangers are stable in all commonly used, aqueous buffers in the range of pH 3–12 and compatible with urea and other strong dissociating agents.
Avoid oxidizing agents and anionic detergents.
Storage
Mono P 5/50 GL, Mono P 5/200 GL, PBE 94, PBE pH 8–10.5
If the column is to be stored for more than two days after use, wash with 5 column volumes of distilled water and 5 column volumes of 20% ethanol.
Polybuffer 96, Polybuffer 74, Pharmalyte 118
Store at 3–8 °C in the dark, preferably under nitrogen. Avoid microbial contamination.
To minimize adsorption of CO2, store solutions under nitrogen in tightly sealed bottles after use.
Never store chromatofocusing media in 1 M HCl or 1 M NaOH. Avoid solutions containing charged groups.
Chromatofocusing and CIPP
Chromatofocusing can give high resolution of quite complex mixtures, but best results will be obtained when working with samples containing few components.
Chromatofocusing is therefore best suited as a polishing step in a purification strategy (Figure 85) when high resolution of similar components is required and the majority of contaminants have been removed.
Pour continuer à lire, veuillez vous connecter à votre compte ou en créer un.
Vous n'avez pas de compte ?