SAE0045
HRV-3C Protease.
N-Terminal His tagged recombinant protein, aqueous solution, 0.8-1.2 mg/mL
Synonym(s):
Human Rhinovirus 3C Protease, Levlfqgp site protease, PreScission Protease
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
UNSPSC Code:
12352204
NACRES:
NA.54
Recommended Products
biological source
human (human Rhinovirus Type 14)
Quality Level
Assay
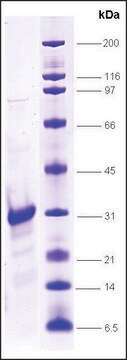
≥90% (SDS-PAGE)
form
aqueous solution
specific activity
≥5000 U/mg
mol wt
21 kDa
concentration
0.8-1.2 mg/mL
technique(s)
protein purification: suitable
suitability
suitable for protein modification
application(s)
life science and biopharma
shipped in
dry ice
storage temp.
−20°C
General description
HRV-3C protease from human rhinovirus type 14 is a protease that specifically cleaves within an eight-residue recognition sequence.
Proteolytic cleavage occurs between the Gln and Gly residues.
HRV-3C protease is a useful tool to cleave recombinant proteins that are expressed as a fusion protein with this sequence, between the carrier domain and the protein of interest.
This recombinant version contains a six-histidine tag and can be easily removed by IMAC chromatography.
Proteolytic cleavage occurs between the Gln and Gly residues.
HRV-3C protease is a useful tool to cleave recombinant proteins that are expressed as a fusion protein with this sequence, between the carrier domain and the protein of interest.
This recombinant version contains a six-histidine tag and can be easily removed by IMAC chromatography.
Application
HRV-3C Protease has been used to remove the GST tag from the DsCystatin protein.
Unit Definition
One unit of HRV-3C Protease is defined as the amount of enzyme needed to digest 1nmole of the substrate peptide per hour (H-Glu-Ala-Leu-Phe-Gln-pNA) at 0 °C, in a reaction buffer consisting of 25 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EDTA, and 1 mM DTT.
Physical form
Supplied in a solution containing 50 mM Tris HCl (pH 7.5), 0.15 M NaCl, 1 mM TCEP and 50% (V/V) glycerol.
Storage Class Code
10 - Combustible liquids
WGK
WGK 2
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Micah Rapp et al.
Cell reports, 35(1), 108950-108950 (2021-04-02)
Antibodies with heavy chains that derive from the VH1-2 gene constitute some of the most potent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-neutralizing antibodies yet identified. To provide insight into whether these genetic similarities inform common modes of recognition, we
An immunosuppressive tick salivary gland protein DsCystatin interferes with Toll-like receptor signaling by downregulating TRAF6
Ta S et al.9
Frontiers in Immunology, 9 (2018)
Richard W Birkinshaw et al.
Molecular cell, 81(10), 2123-2134 (2021-04-02)
A body of data supports the existence of core (α2-α5) dimers of BAK and BAX in the oligomeric, membrane-perturbing conformation of these essential apoptotic effector molecules. Molecular structures for these dimers have only been captured for truncated constructs encompassing the
Zhengmao Xu et al.
Parasites & vectors, 12(1), 341-341 (2019-07-13)
Rhipicephalus haemaphysaloides is a widespread tick species in China and other South East Asian countries, where it is the vector of many pathogens. The objective of this study was to study the role of serpin (serine protease inhibitor) during the
Zhonggang Hou et al.
Current protocols, 2(2), e361-e361 (2022-02-08)
CRISPR-Cas systems provide researchers with eukaryotic genome editing tools and therapeutic platforms that make it possible to target disease mutations in somatic organs. Most of these tools employ Type II (e.g., Cas9) or Type V (e.g., Cas12a) CRISPR enzymes to
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service