Manufacturing Strategies for mRNA Vaccines and Therapeutics
Laurens Vergauwen, Process Development Scientist, Nargisse El Hajjami, Associate Director Cell and Gene Therapy Segment, Manuel Brantner, Associate Director Vaccine and Plasma Segment, Shiksha Mantri, Global Marketing Manager mRNA Applications, Bahar Cebi, Segment Marketing Manager Novel Modalities & Vaccines
Merck
Viral delivery systems such as adeno-associated virus (AAV) vectors are well established and approved for vector immunogenicity use as vaccines and gene therapies. Despite their widespread use, viral delivery systems can lead to immunogenicity and more frequent systemic side effects than other modalities. In addition, the manufacturing process can be complex and high titers are needed for gene therapy applications.
In contrast, non-viral delivery systems are expected to have a better safety profile and with simplified manufacturing, offer the potential for a templated process.
mRNA technology uses non-viral delivery systems and offers a great deal of versatility. Delivery of an mRNA into the cytosol of a cell can induce the production of a target protein which can function as a therapeutic or prophylactic, act as an antigen to trigger an immune response for vaccination purposes, replace a defective protein or activate an anti-tumor response.
Strong early results for three mRNA vaccines against SARS-CoV-2 have implications that go far beyond the current pandemic and bode well for similar approaches in the fight against cancer, heart disease and other infectious diseases.
View all of our products and services for mRNA development and manufacturing.
Considerations for mRNA Manufacturing
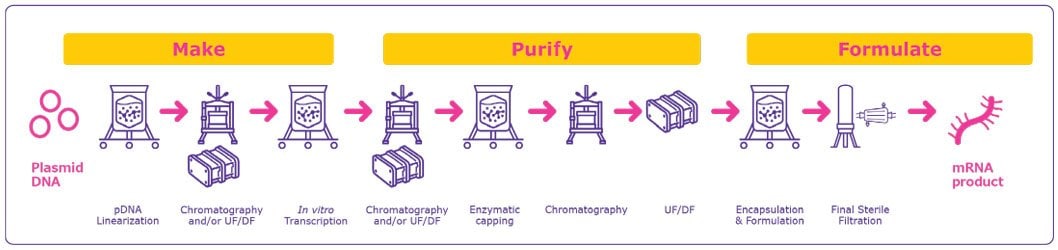
The development and manufacturing of mRNA for use as therapeutics and vaccines are comparatively simple, scalable and extremely rapid (Figure 1). With a compressed timeframe from development to clinic and towards approval, mRNA technology is attractive not only for response to outbreaks of infectious diseases and pandemics, but also for development of novel therapeutic approaches for addressing diseases with unmet needs.
mRNA is produced by in vitro synthesis through an enzymatic process; this contrasts with classical in vivo protein expression where time-consuming cloning and amplification steps are needed. Because an in vitro synthesis process is used, there is no need to remove cells or host cell proteins. This simplified manufacturing process, which uses the same reaction materials and vessels for any target, allows GMP facilities to switch to a new protein target within a very short period of time, with minimal adaptation to process and formulation.
Find detailed information about products, services and expertise by downloading our brochure: "Enabling Capabilities and Solutions for all mRNA (Nucleic Acid) platforms"
Making the mRNA
Production of mRNA-based therapeutics and vaccines typically or commonly begins with a pDNA template that contains a DNA-dependent RNA polymerase promoter and the corresponding sequence for the mRNA construct. Given the central role of the pDNA construct, its design and purity are important factors for optimizing the mRNA product. pDNA production and purification present several challenges due to the large size of the nucleic acid and its high viscosity, shear sensitivity and the similarities between the pDNA and impurities. Strategies to overcome these challenges are addressed in our webinar entitled “Scalable purification of Plasmid DNA”.
The mRNA construct is designed to ensure efficient expression of the gene of interest. Stability, gene expression and efficient protein translation depend upon several structural elements (Figure 2):
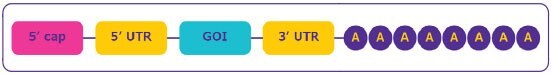
Figure 2.mRNA structure.
- The cap region at the 5’ end of the sequence is essential for mRNA maturation and allows the ribosome to recognize the mRNA for the efficient protein translation. The cap also stabilizes mRNA by protecting it from nuclease digestion.
- The untranslated regions (UTRs) located at the upstream and downstream domains of the mRNA coding region are affecting translation efficiency, localization and stability and can be utilized for efficient protein expression.
- The open reading frame or coding sequence regions contains the gene of interest (GOI).
- The poly-(A) tail is crucial for protein translation and mRNA stability by preventing digestion by 3’ exonuclease.
The required pDNA is amplified within bacterial cells, typically E. coli, and subsequent purification steps yields a pure, concentrated, circular pDNA. The pDNA is then linearized to serve as a template for the RNA polymerase to transcribe the desired mRNA.
Linearization is required to avoid transcriptional readthrough events that may generate undesired forms of mRNAs leading to additional impurities that would need to be removed. Linearization is achieved by mixing the plasmid DNA with a restriction enzyme in a reaction buffer4 and subsequent incubation at 37 °C for 4 hours. Optionally, the reaction is stopped by the addition of EDTA or heat inactivation at 65 °C.
Impurities such as the restriction enzyme, BSA, DNA fragments, endotoxins and others are then removed. Most of the lab scale processes use a solvent extraction technique and this not applicable for GMP production environments.
As an alternative, tangential flow filtration (TFF) and chromatography are efficient impurity removal techniques for this purification step.
The next step is in vitro transcription during which the linearized pDNA, serving as the DNA template, is transcribed into mRNA. This enzymatic reaction uses elements of the natural transcription process, including RNA polymerase and nucleotide triphosphates. Following transcription, the final mRNA structure requires a 5’ cap structure for stability and efficient transduction in the cell.
The cap can be added in two ways – either co-transcriptionally or enzymatically. Co-transcriptional capping is usually accomplished by adding cap analogs and guanosine triphosphate (GTP) in the transcription mix at a ratio of four cap analogs for one GTP. Following an incubation step at 37 °C, the DNA template is typically degraded by the addition of DNases; the resulting small DNA fragments can then be easily separated from larger mRNA molecules by tangential flow filtration (TFF). Another option to remove the DNA template includes the utilisation of a chrome step (e.g. Poly (dT) capture). In the latter case the DNA template does not need to be digested, which avoids the risk of small DNA fragments hybridizing to the mRNA.4
Co-transcriptional capping is less expensive and faster than enzymatic capping as it is performed during the transcription step, in the same reactor mix. However, efficiency and yield are lower and it can generate non-capped impurities as GTP can bind to the mRNA sequence instead of the cap analogs. In addition, the cap analogs can be incorporated in the reverse orientation. To overcome this, some antireverse cap analogs (ARCA) have been developed to prevent this reverse incorporation of a 5’ cap, leading to higher translation efficiency.
Enzymatic capping is performed after mRNA purification from the in vitro transcription mixture. This reaction usually uses a vaccinia virus-capping enzyme to add the capping structure to the mRNA structure. While enzymatic capping has a very high capping efficiency, it is more expensive and requires an extra unit operation.
Purifying mRNA
Following the in vitro transcription step, mRNA is purified from the impurities and materials used in the previous steps including endotoxins, immunogenic double stranded RNA (dsRNA), residual DNA template, RNA polymerase and elemental impurities. Several options are available for mRNA purification.
TFF allows efficient separation of mRNA from smaller impurities that are not retained by the membrane; molecular weight cut-offs ranging from 30 to 300 kDa can be used based on the size of the mRNA. With TFF it is possible to purify, concentrate and diafilter the product within the same unit operation. At this stage, the mRNA will need to be in the appropriate buffer, either for enzymatic capping or chromatography. An important consideration when using TFF, however, is that small DNA fragments can hybridize to the mRNA, generating additional impurities. As seen before, you don't have this risk if using capture to remove DNA template.5
A number of chromatography techniques can be used as an alternative to TFF and include reverse-phase ion pair, anion exchange and affinity chromatography using poly(dT) capture (Table 1).

Table 1.Comparison of reversed-phase ion-pair, anion exchange and affinity chromatography for mRNA purification. DBC: dynamic binding capacity.3,4
Chromatography provides an efficient means for DNA template removal and eliminates the risk of hybridization that can occur during Ultra-/Diafiltration step. It is, however, more expensive and a TFF step would still be required for media exchange and preparation for the subsequent step.
Chromatography is also used following the enzymatic capping step to remove unwanted products and oligonucleotide impurities coming from the previous enzymatic reaction steps.
Reversed-phase ion pairing is commonly used at small scales and allows a very efficient and rapid RNA purification and good separation of single stranded RNA (ssRNA) from DNA, double stranded RNA (dsRNA), and short transcripts. However, this method uses solvents making it poorly suitable for GMP manufacturing production. The technique also requires ion-pair reagents and resulting formation of complexes with the mRNA may require extensive diafiltration steps for removal. Furthermore, its sensitivity to fouling by proteins and aggregates makes this technique better suited for polishing than for capture.
Anion exchange has a high dynamic binding capacity and is very efficient for removing immunogenic impurities such as dsRNA, uncapped RNA, RNA–DNA hybrids and other RNA structures. While this allows the use of aqueous solutions, it might require the addition of chaotropic agents that can be toxic and operation at temperatures of up to 85°C to desorb large mRNA molecules bound to the resin. Ambient temperature operations typically elute mRNA species smaller than 500 bases.3
Affinity chromatography poly(dT) capture uses a resin to specifically capture the poly(A) tail of full-length mRNA transcripts. This process efficiently removes DNA, nucleotides, enzymes, buffer components and any other impurities not having a poly(A) tail.
The downside of this technique is that, unlike reversed phase and anion exchange, it cannot discriminate dsRNA from ssRNA. In addition, product-related poly(dT) is not efficient for removing other product-related impurities such as DNA fragments that have hybridized to the mRNA. For this reason, the initial chromatography step is Affinity Chromatography typically followed by a second chromatography step using anion exchange for polishing purposes.
Following the chromatography step(s), a final concentration and diafiltration is performed to maximize product purity and transfer the mRNA into the appropriate buffer for formulation or storage. At this stage, mRNA can be further purified, concentrated and diafiltered within the same unit operation. A sterile filtration step can be performed following this TFF step. It should be noted, however, that sterilizing grade filtration of some mRNAs with a molecular weight of 5000 kDa or higher can be challenging.
Formulating the mRNA
Delivery tools are equally important in the effectiveness of mRNA vaccines and therapeutics. After the final mRNA purification step, the next consideration is the delivery mechanism (Figure 3). One of the most advanced class of delivery systems are combinations of lipids and polymers. These include complexes of oligonucleotides bound to lipids forming a lipoplex or positively charged polymers such as polyethyleneimine (PEI) forming polyplexes.
Lipid nanoparticles (LNP) are most commonly used for mRNA delivery; each lipid nanoparticle consists of four different lipids allowing the mRNA to be carried in it and protected from degradation.
Cationic/ionizable lipids are required for encapsulating the RNA via electrostatic interactions. Delivery to hepatocytes (for boosting or silencing of protein expression) requires ionizable lipids (passive targeting, endosomal release) whereas uptake by immune cells is much easier. It also works with strong cationic lipids.

Figure 3.Several mRNA delivery systems are available.
These lipids are also responsible for efficient release of the RNA into the cytoplasm. The structure of cationic lipids has a major impact on the activity of the LNP, its toxicity and biodistribution, which then influences potential toxicity effects in the body.
Polyethylene glycol (PEG) lipids provide colloidal stability and prevent protein binding to the particle, thereby shielding it from the immune system and achieving longer circulation. The length of the PEG chain and fatty acid chains determine the circulation lifetime and fusogenicity, or how well the particle can fuse with the endosomal membrane of the LNP. If the goal is prolonged circulation, longer fatty acid chains can be used, such as polyethylene glycoldistearoylglycerol (DSG PEG 2000). The concentration of PEG also has an effect on the size of the particle. In addition, use of PEG may result in the formation of antibodies against it, potentially rendering the immunization useless.
Neutral/anionic lipids provide structural stability and play a role in defining the fusogenicity and biodistribution. 1,2-dioleoyl-sn-glycero-3- phosphoethanolamine. For example, a recent study1 showed LNPs containing 1,2-dioleoyl-sn-glycero- 3-phosphoethanolamine (DOPE), which plays an important role in endosomal release, led to enhanced delivery of mRNA to the liver as compared to 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC). Recent studies2 suggest that these helper lipids also assist in the stable encapsulation of the RNA.
Cholesterol is used to modulate the bilayer density, fluidity and uptake (raft formation) of the LNP. While there are animal-derived and synthetic versions of cholesterol available in the market, synthetic cholesterol offers several advantages including higher purity, lack of animal derived molecules such as prions, scalability, and highly consistent quality.
Considerations for lipid selection Lipids should be chosen based on the delivery route in mind to achieve maximum efficacy and optimal biodistribution. In addition to the choice of lipids, the ratio between the individual lipids it is an important component to finetune, as it has a direct impact on the bilayer fluidity and the fusogenicity of the LNP.
Several critical aspects must be considered when selecting the lipid. Lipid type, source and quality have a direct impact on the impurity profile and properties such as the particle characteristics, stability and release profile is the final formulation. To achieve reproducible results with the final formulation, consistent quality of lipids is required, which is dependent on the quality of the raw materials used to synthesize the lipids and appropriate material characteristics of the lipid itself.
The purified mRNA can be formulated into the delivery particle via different techniques. In the commonly used solvent injection technique, lipids are dissolved in a solvent such as ethanol and rapidly mixed in an aqueous, low pH buffer containing the mRNA using a crossflow mixing or microfluidic mixing is to create the LNPs. The low pH buffer is then diafiltered into a neutral buffer and ultrafiltration is used to concentrate the particles.
The TFF step must be rapid as lipids can be hydrolyzed at low pH, leading to formation of impurities such as hydrolipids that can affect the lipid bilayer structure, stability of the formulation and drug release characteristics. Degradation of the lipids can also increase the size of the particle, resulting in aggregation.
LNPs have a very good stability, structural plasticity and enhanced gene delivery compared to other delivery systems. They increase the transfection rate compared to naked mRNA, allow for intravenous injection without the risk of being degraded by RNases present in the bloodstream and enable active targeting if specific ligands are incorporated.
Disadvantages of LNPs include the fact that they may require cold chain logistics. In addition, sterile filtration is not always possible with LNPs and in such cases alternatives, such as gamma irradiation, heat sterilization, high-pressure sterilization or closed processing must be considered.
For a more detailed discussion of mRNA formulation and its related critical parameters and considerations, please view our webinar: "Key to Successful Formulation Development for Lipid Based RNA Delivery".
Scale-up Considerations
There are several considerations to keep in mind when scaling up the mRNA manufacturing process, and these should be top-of-mind during process development when working in a small scale.
- Methods using solvent extraction and precipitation steps for mRNA purification are difficult to scale and the use of hazardous solvents are not suitable for GMP environments and can be replaced by TFF or chromatography.
- Because mRNA can be degraded by RNases within seconds, every raw material, solution and equipment that comes into contact with the product must be free of these enzymes.
- The appropriate delivery system contributes to the efficiency of the vaccine or therapeutic and should be selected carefully.
- If the final product is a large mRNA complex, alternatives for the sterile filtration of the product should be evaluated.
- Extraordinary supply chain requirements (e.g. cold chain) are a significant cost driver. Therefore, the stability of the drug should be evaluated carefully.
A Bright Future
mRNA technology has enabled development of COVID-19 vaccine candidates with unprecedented speed and outstanding efficacy rates. Going forward, this technology will not only revolutionize the field of vaccine development by allowing a rapid response to disease outbreaks, it will also help to address diseases of unmet medical need with gene therapy approaches.
mRNA has the potential to be a rapid and flexible vaccine platform; vaccine development can now focus on process development, rather than being a scientific challenge. Companies can design mRNA vaccines relatively quickly once they know the genetic sequence of the pathogen. To ensure this therapeutic approach reaches its full potential, however, innovative solutions, expertise and ingenuity will need to coalesce to establish a simple and robust platform at production scale. Concerns related to safety, efficacy, quality and manufacturability of the developed mRNA constructs and the delivery systems will also need to be evaluated.
Once these challenges are addressed, mRNA-based vaccines and therapeutics will likely take their place among the advanced modalities delivering remarkable results for patients around the world.
References
Um weiterzulesen, melden Sie sich bitte an oder erstellen ein Konto.
Sie haben kein Konto?