SML1697
OGG1 Inhibitor O8
≥98% (HPLC)
Sinónimos:
3,4-Dichloro-benzo[b]thiophene-2-carboxylic acid hydrazide
About This Item
Productos recomendados
Quality Level
assay
≥98% (HPLC)
form
powder
color
white to beige
solubility
DMSO: 5 mg/mL, clear
storage temp.
2-8°C
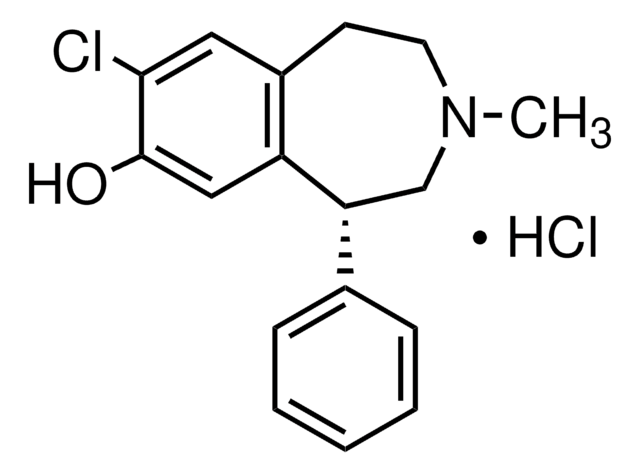
SMILES string
ClC1=C(C(NN)=O)SC2=CC=CC(Cl)=C21
InChI
1S/C9H6Cl2N2OS/c10-4-2-1-3-5-6(4)7(11)8(15-5)9(14)13-12/h1-3H,12H2,(H,13,14)
InChI key
HSSHUDKWJRJKPV-UHFFFAOYSA-N
General description
Biochem/physiol Actions
signalword
Warning
hcodes
Hazard Classifications
Eye Irrit. 2 - Skin Irrit. 2 - STOT SE 3
target_organs
Respiratory system
Storage Class
11 - Combustible Solids
wgk_germany
WGK 3
flash_point_f
Not applicable
flash_point_c
Not applicable
Elija entre una de las versiones más recientes:
Certificados de análisis (COA)
¿No ve la versión correcta?
Si necesita una versión concreta, puede buscar un certificado específico por el número de lote.
¿Ya tiene este producto?
Encuentre la documentación para los productos que ha comprado recientemente en la Biblioteca de documentos.
Nuestro equipo de científicos tiene experiencia en todas las áreas de investigación: Ciencias de la vida, Ciencia de los materiales, Síntesis química, Cromatografía, Analítica y muchas otras.
Póngase en contacto con el Servicio técnico