SEQRI
SEQPLEX-I WTA Kit
Whole Transcriptome Amplification, RNA Amplification
Synonym(s):
RNA amplification kit
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
UNSPSC Code:
41121800
NACRES:
NA.55
Recommended Products
technique(s)
whole genome amplification: suitable
whole transcriptome amplification: suitable
compatibility
Illumina (Next Generationa Sequencing)
shipped in
wet ice
storage temp.
−20°C
General description
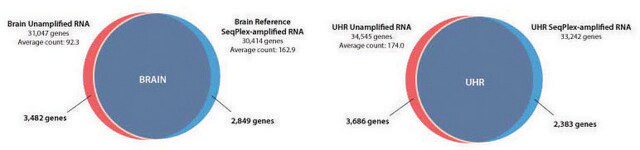
The SeqPlex™-I whole transcriptome amplification (WTA) kit allows the amplification of small quantities of reverse transcribed RNA or degraded RNA for direct input onto Illumina® next-generation sequencing (NGS) flow cells. The SeqPlex-i process is comprised of three steps:
Pre-amplification/Library Synthesis: In the Pre-amplification/Library Synthesis step using the (Library Preparation Reagents), the template RNA is reverse transcribed using primers composed of a semi-degenerate 3′- and universal 5′-ends. As polymerization proceeds, displaced and RNaseH generated single strands serve as new templates for additional primer annealing and extension producing random, overlapping cDNAs flanked by a universal primer (5′) and primer complement (3′) sequence.
Amplification 1: In the Amplified Library Synthesis step (using the Amplification 1 Reagents), products from pre-amplification/library synthesis are amplified by single primer PCR via the proprietary universal end sequence. These amplification products typically range from 200 to 500+ base pairs. Note: Amplicons from degraded RNA, such as Formalin Fixed Paraffin Embedded (FFPE), are typically shorter and dependent upon the length of the starting RNA.
Amplification 2: In the Sequencing Library Synthesis step (using Amplification 2 Reagents), single primer amplicons from amplification 1 are converted to dual Illumina® primer PCR products ready for purification, quantification, and Illumina® NGS.
Pre-amplification/Library Synthesis: In the Pre-amplification/Library Synthesis step using the (Library Preparation Reagents), the template RNA is reverse transcribed using primers composed of a semi-degenerate 3′- and universal 5′-ends. As polymerization proceeds, displaced and RNaseH generated single strands serve as new templates for additional primer annealing and extension producing random, overlapping cDNAs flanked by a universal primer (5′) and primer complement (3′) sequence.
Amplification 1: In the Amplified Library Synthesis step (using the Amplification 1 Reagents), products from pre-amplification/library synthesis are amplified by single primer PCR via the proprietary universal end sequence. These amplification products typically range from 200 to 500+ base pairs. Note: Amplicons from degraded RNA, such as Formalin Fixed Paraffin Embedded (FFPE), are typically shorter and dependent upon the length of the starting RNA.
Amplification 2: In the Sequencing Library Synthesis step (using Amplification 2 Reagents), single primer amplicons from amplification 1 are converted to dual Illumina® primer PCR products ready for purification, quantification, and Illumina® NGS.
To order SeqPlexI adapters in tubes, please download NGSO Adapters SeqPlexI tubes (XLSX File). To order arrayed adapters in plates, download NGSO Plate SeqPlexI arrayed plates (XLSX File). Details on our custom adapters product, Next-Gen Sequencing Oligos (NGSO), can be found at SigmaAldrich.com/nextgenoligos. From this page, you can directly access an online ordering configurator by clicking on the "Order Now" option. Both spreadsheets are directly uploadable after selecting the quantity and/or format from the dropdown menus. For adapters in solution in tubes or in plates, NGSO-Silver is the only option available, which has a length cutoff of 60 bases (several of the adapters are longer than 60 bases). In addition, adapters in plates cannot be duplexed. If you would like a feasibility assessment of ordering NGSO-Silver longer 60 bases or duplexed in plates, then please send a request to dnaoligos@milliporesigma.com. All of the adapters are readily available for online ordering if dry and in tubes as either NGSO-Bronze or Gold (recommended purification for any adapter).
Application
SeqPlex™-I WTA Kit has been used for whole transcriptome amplification. It has also been used to sequence the total nucleic acid from chikungunya virus (CHIKV) cytopathic effect (CPE) positive cell culture samples.
Features and Benefits
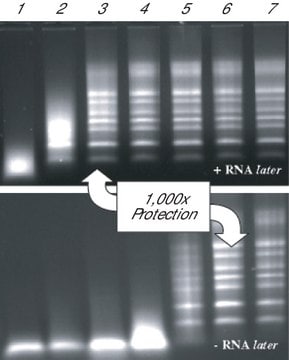
- Amplifies fragmented/extremely small quantities of total RNA: Fragmented or intact RNA from all sources including FFPE and RIP are easily amplified by random priming technology.
- Semi-degenerate library primer design ensures more complete transcriptome coverage and efficient priming
- Fewer Steps: No need to fragment cDNA before sequencing
- High-efficiency: Amplifies ds-cDNA in 8 hours or less
- Cost-effective: No longer requires an additional NGS library prep step
- Compatible with Illumina® next generation sequencing
Other Notes
1) RNA Handling Technique
a) The reagents in this kit have been tested to assure that RNases are absent.
b) The user, however, must protect the integrity of experimental results by wearing basic protective equipment, including gloved hands and lab coat.
c) All reagent transfers throughout this procedure should be performed in a laminar flow hood or dedicated clean room.
d) Frozen RNA samples should be thawed on ice.
2) A 20 μL Amplification 2 reaction will produce >100 ng of amplified double-stranded cDNA when starting with 100 pg to 5 ng of high-quality RNA. Higher input quantities and higher quality RNA template generally result in increased yields. For damaged RNA, such as from FFPE, 1–50 ng input RNA is recommended.
3) The dual index adapter primers (AP100) provided in this kit will only work for one sample. If pooling samples for sequencing is required, the user must provide additional index primer sets. See example index primer sequences on page 2 of the technical bulletin.
a) The reagents in this kit have been tested to assure that RNases are absent.
b) The user, however, must protect the integrity of experimental results by wearing basic protective equipment, including gloved hands and lab coat.
c) All reagent transfers throughout this procedure should be performed in a laminar flow hood or dedicated clean room.
d) Frozen RNA samples should be thawed on ice.
2) A 20 μL Amplification 2 reaction will produce >100 ng of amplified double-stranded cDNA when starting with 100 pg to 5 ng of high-quality RNA. Higher input quantities and higher quality RNA template generally result in increased yields. For damaged RNA, such as from FFPE, 1–50 ng input RNA is recommended.
3) The dual index adapter primers (AP100) provided in this kit will only work for one sample. If pooling samples for sequencing is required, the user must provide additional index primer sets. See example index primer sequences on page 2 of the technical bulletin.
Legal Information
Illumina is a registered trademark of Illumina, Inc.
SeqPlex is a trademark of Sigma-Aldrich Co. LLC
Disclaimer
The SeqPlex-I DNA Amplification Kit for whole genome amplification (WGA) is for R&D use only. Not for drug, household, or other uses.
Signal Word
Danger
Hazard Statements
Precautionary Statements
Hazard Classifications
Resp. Sens. 1
Storage Class Code
10 - Combustible liquids
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
John T Kayiwa et al.
Tropical diseases, travel medicine and vaccines, 5, 21-21 (2019-12-05)
Arboviruses are (re-) emerging viruses that cause significant morbidity globally. Clinical manifestations usually consist of a non-specific febrile illness that may be accompanied by rash, arthralgia and arthritis and/or with neurological or hemorrhagic syndromes. The broad range of differential diagnoses
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service