913812
2-(Azidomethyl)nicotinic acid imidazolide
≥95%
Synonym(s):
2-(Azidomethyl)-3-(1H-imidazole-1-carbonyl)pyridine, NAI-N3, RNA SHAPE probe, icSHAPE reagent
About This Item
Recommended Products
Quality Level
Assay
≥95%
form
powder
mp
74-80 °C
storage temp.
−20°C
SMILES string
[N+](=[N-])=NCc1ncccc1C(=O)[n]2cncc2
InChI
1S/C10H8N6O/c11-15-14-6-9-8(2-1-3-13-9)10(17)16-5-4-12-7-16/h1-5,7H,6H2
InChI key
QPSQVTFTPHUCEH-UHFFFAOYSA-N
Application
Other Notes
Signal Word
Danger
Hazard Statements
Precautionary Statements
Hazard Classifications
Eye Irrit. 2 - Self-react. C - Skin Irrit. 2
Storage Class Code
5.2 - Organic peroxides and self-reacting hazardous materials
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Choose from one of the most recent versions:
Certificates of Analysis (COA)
It looks like we've run into a problem, but you can still download Certificates of Analysis from our Documents section.
If you need assistance, please contact Customer Support.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service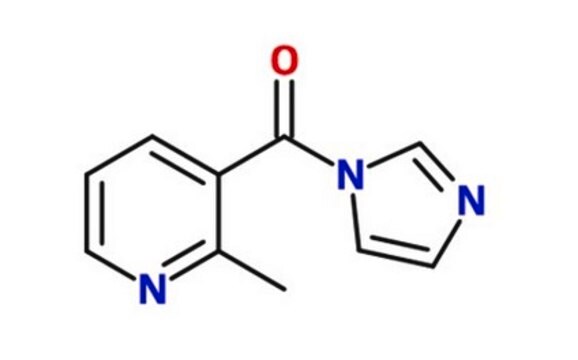
![Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine 97%](/deepweb/assets/sigmaaldrich/product/structures/179/695/86a721c8-2a4c-4e4f-bc36-6276ce7a941f/640/86a721c8-2a4c-4e4f-bc36-6276ce7a941f.png)