Degrader Building Blocks for Targeted Protein Degradation
Targeted Protein Degradation
An emerging strategy in drug discovery to access difficult-to-treat diseases is targeted protein degradation. While traditional small-molecule or antibody drugs may only allow access to ~20% of the proteome, degradation techniques may open the door to the other 80%.1 The molecules used in these approaches are often called protein degraders (such as proteolysis-targeting chimeras, or PROTAC® molecules), which are bifunctional molecules that eliminate target proteins from cells (Figure 1).1-5
*PROTAC® is a registered trademark of Arvinas Operations, Inc., and is used under license.
Targeted protein degradation via proteolysis-targeting chimeras (PROTACs)
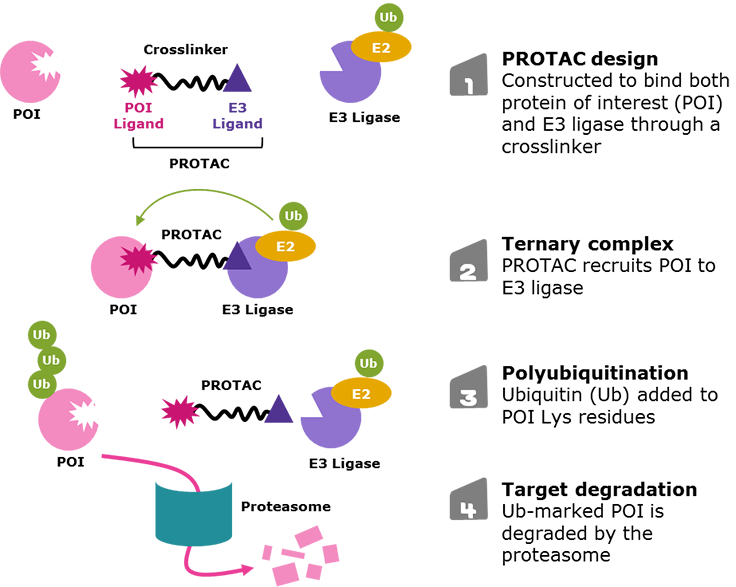
Figure 1.Targeted protein degradation via proteolysis-targeting chimeras (PROTACs)
PROTACs are designed with three primary components:
- A ligand at one end that targets the protein of interest (POI)
- A second ligand at the opposite end that binds an E3 ligase
- A crosslinker in the middle that joins the two ends (Figure 2).
The simultaneous PROTAC binding of two proteins brings the POI in close enough proximity for polyubiquitination by the E2 enzyme associated to the E3 ligase, which flags the POI for degradation through the proteasome.1-5
Learn more from Professor Alessio Ciulli in our recent webinar with the Royal Society of Chemistry: Targeted protein degradation using small molecules.
Degrader Building Blocks for Protein Degradation and PROTAC Research
The design of small molecules for target degradation is not trivial since even slight alterations in ligands and crosslinkers can affect binding to the POI or E3 ligase or the formation of the ternary complex.3-5 Thus, many analogs are synthesized – varying each structure slightly – and screened in cells to discover the optimal PROTAC or degrader for target degradation. To streamline this synthesis, our protein degrader building blocks are a collection of crosslinker-E3 ligase ligand conjugates with a pendant functional group for covalent linkage to a target ligand (Figure 2). Furthermore, because the same functional group is present across a series, one target ligand can be conjugated to several of our protein degrader building blocks in parallel for facile library generation and subsequent screening (Figure 3).
Degrader building blocks are permutations of the following components:
- Ligands targeting the E3 ligase Cereblon (CRBN) or von Hippel–Lindau (VHL)
- Crosslinkers with varied lengths and compositions
- Conjugation sites with reactivity for common functional groups
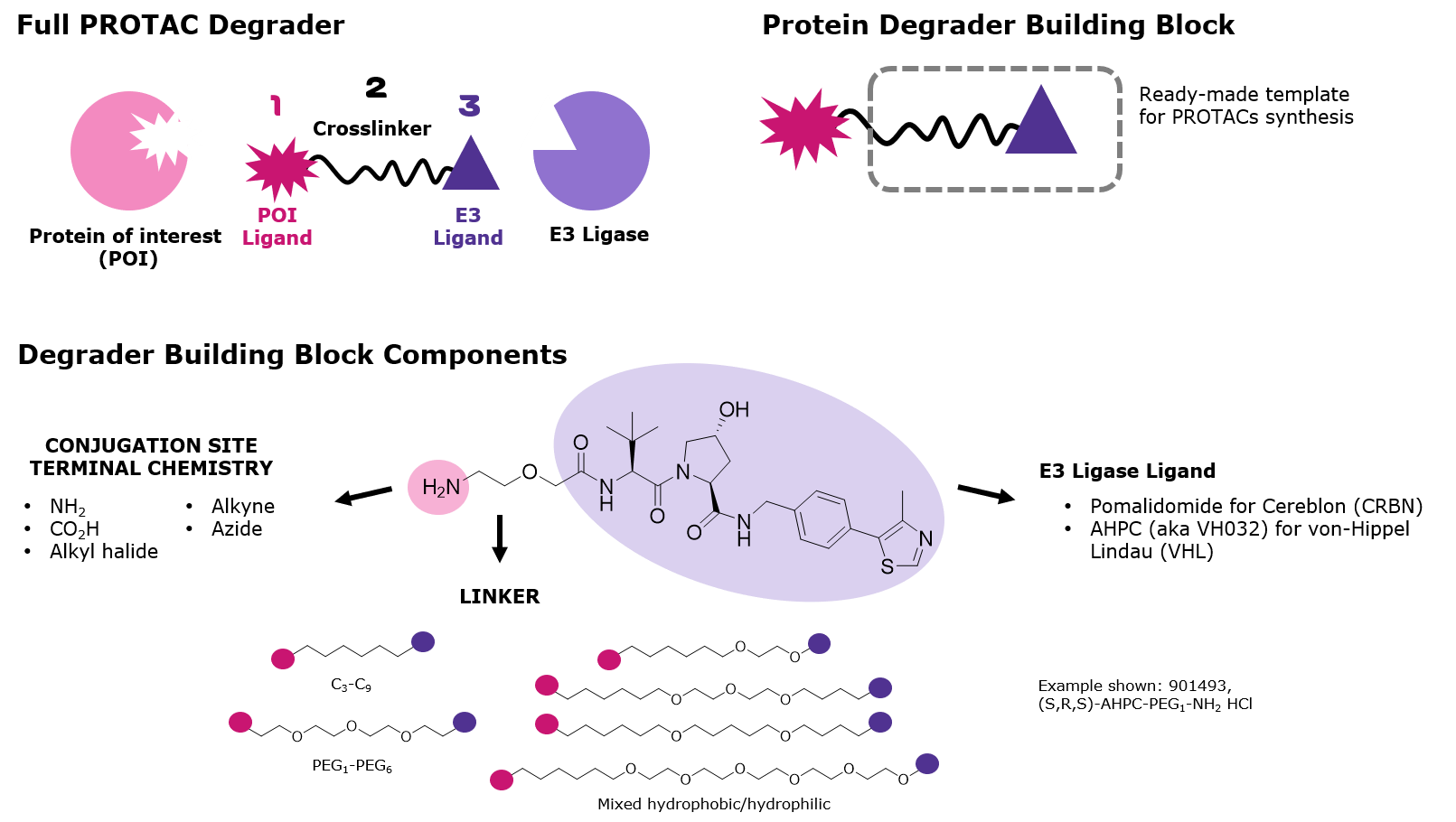
Figure 2.Primary components of PROTAC protein degraders
Advantages
- Compatibility: Linkers conjugate to common functional groups present on target ligands.
- Molecule design: Strategic variety encompassed in the combinations of linkers and ligands aids the design of target degraders.
- Synthetic time-saver: The E3 ligand-crosslinker conjugates decrease the amount of time spent on PROTACs synthesis.
- Library generation: Using degrader building blocks with the same conjugation site enables the simultaneous generation of several protein degraders via parallel synthesis.
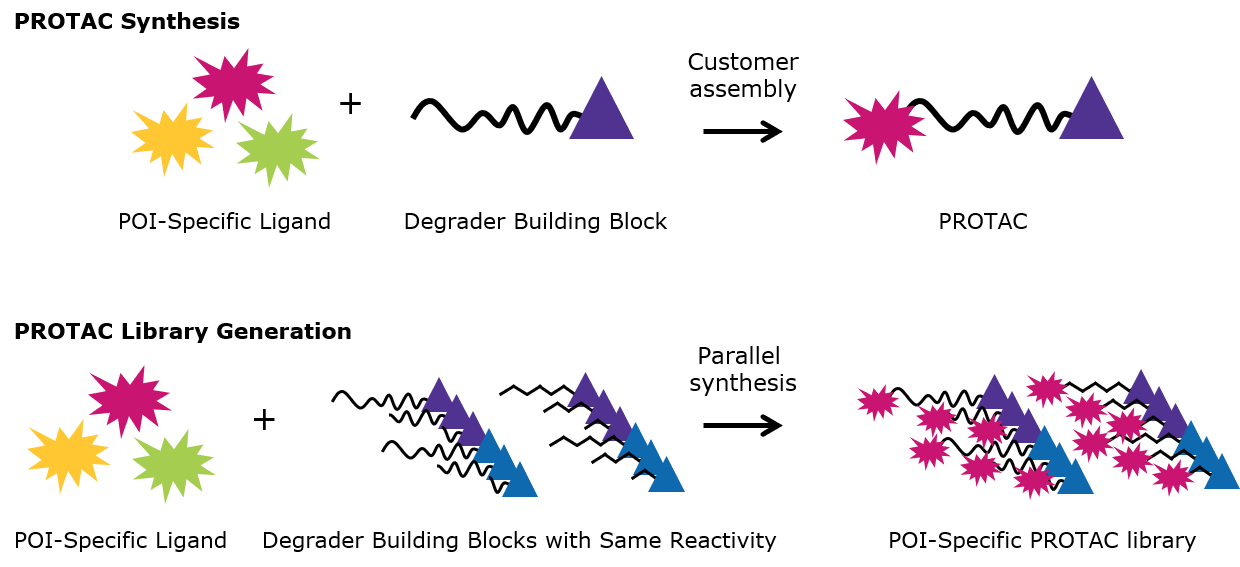
Figure 3.PROTAC Synthesis and PROTAC Library Generation
| Degrader building blocks simplify the synthesis of PROTAC protein degraders. |
| Use of a set of degrader building blocks with the same conjugation site streamlines the synthesis of libraries. |
Please contact us for more information, bulk inquiries, or a full ChemDraw/sdf file of protein degrader building blocks and crosslinkers.
References
如要继续阅读,请登录或创建帐户。
暂无帐户?