Kapa Biosystems Taq DNA Polymerase and PCR Kits
Taq DNA polymerase PCR reagents and kits for DNA extraction and amplification
DNA extraction and amplification are critical steps in your molecular biology workflow and often requires purified, high-quality DNA. Kapa Biosystems offers a robust portfolio of DNA polymerase reagents and PCR kits for extracting and amplifying a variety of mammalian and plant-based genetic materials. Incorporating revolutionary thermostable polymerases, our Kapa Biosystems reagents and kits are designed to enable your next discovery with improved sensitivity and specificity for multiple applications.
- KAPA Taq DNA Polymerase
- KAPA2G Fast Multiplex
- KAPA2G Robust DNA Polymerase
- KAPA Long Range PCR System
- KAPA Mouse Genotyping
- KAPA Express Extract
- KAPA3G Plant PCR Kits
KAPA Taq DNA Polymerase
High-quality and performance for routine PCR.
KAPA Taq DNA Polymerase is based on the single-subunit, wild-type Taq DNA polymerase of the thermophilic bacterium Thermus aquaticus. KAPA Taq and KAPA Taq HotStart DNA Polymerase have 5'-3' polymerase and 5'-3' exonuclease activities, but no 3'-5' exonuclease proofreading activity. The enzyme has an error rate of approximately 1 error per 2.2 x 105 nucleotides incorporated.
KAPA Taq DNA Polymerase kits are ideally suited for:
- Standard PCR
- DNA labelling
- DNA sequencing
- Numerous applications for which a high-quality, thermostable DNA polymerase is required
Benefits include:
- Improved sensitivity, specificity, and yields
- Novel buffer formulation facilitates specific primer annealing, leading to higher yield of a specific product
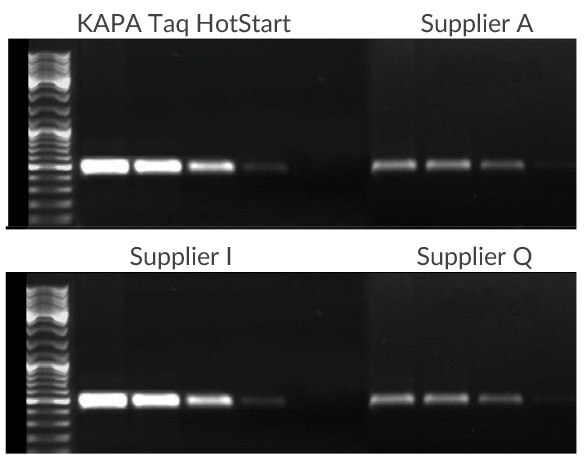
Figure 1.A 500 bp fragment of the CCR5 gene was amplified using 100 ng, 10 ng, 1 ng, or 100 pg of human genomic DNA as template. KAPA Taq HotStart exhibits improved sensitivity, specificity, and yield when compared with competitive HotStart products. All reactions were performed using the manufacturer recommended protocols.
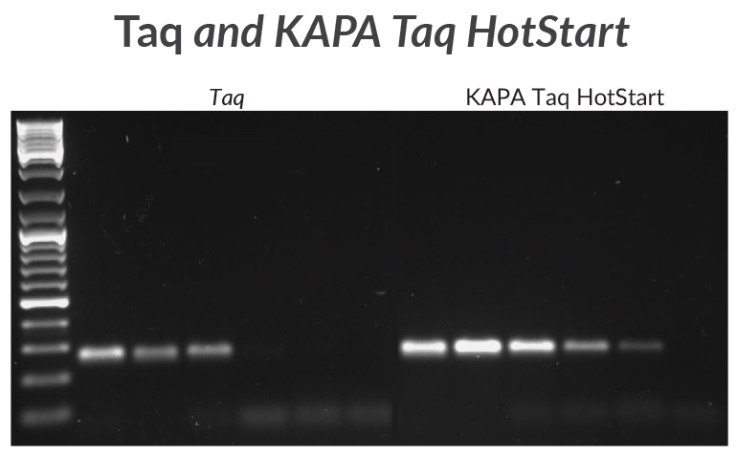
Figure 2.A 270 bp amplicon was amplified from mycoplasma DNA with KAPA Taq or KAPA Taq HotStart. Sensitivity was tested using a 10X template dilution series starting with 1 ng of DNA.
KAPA2G FAST MULTIPLEX
High-speed, high-performance multiplex PCR.
KAPA2G Fast Multiplex PCR Kits contain a second-generation (2G) enzyme derived through a process of directed evolution. KAPA2G FAST HotStart DNA Polymerase is an antibody-mediated hot start formulation engineered for higher processivity and speed, offering significantly faster extension rates than wild-type Taq DNA polymerase. In addition to speed, KAPA2G Fast provides higher yields and sensitivity than competitor enzymes for highly multiplexed PCR.*
KAPA2G FAST Multiplex provides:
- Uniform representation of all amplicons
- Successful multiplex PCR with difficult, GC-rich targets
- Reduction in PCR cycling time up to 60%
- Extension times as low as 15 seconds
- High speed without compromising performance
- Minimal optimization with master mix formulation
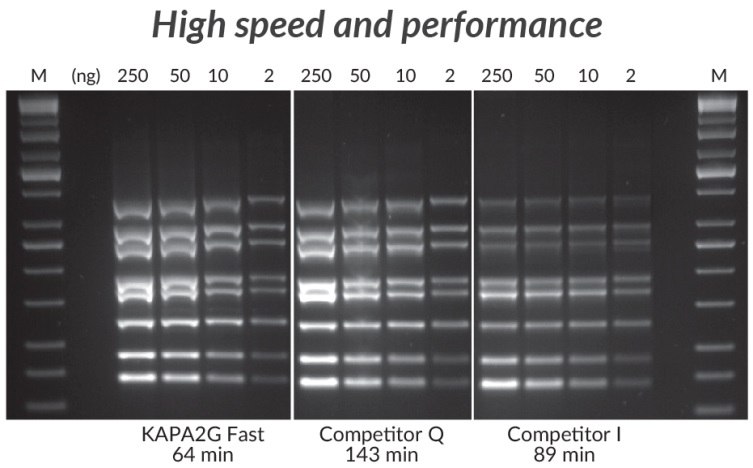
Figure 3 .*Multiplex PCR (8-plex) performed with the KAPA2G Fast Multiplex PCR Kit, Competitor Q, and Competitor I. Reactions (25 μL) contained 1X PCR Master Mix (KAPA and Competitor Q) or 1X PCR Buffer, 3 mM MgCl2, 0.2 mM of each dNTP and 1 U of hot start Taq DNA Polymerase (home-brew multiplex reagents, with Competitor I). Human genomic DNA was used as template (250 – 2 ng per reaction), and primers were supplied at 0.2 μM each. Cycling was performed according to manufacturers’ recommendations (30 cycles).
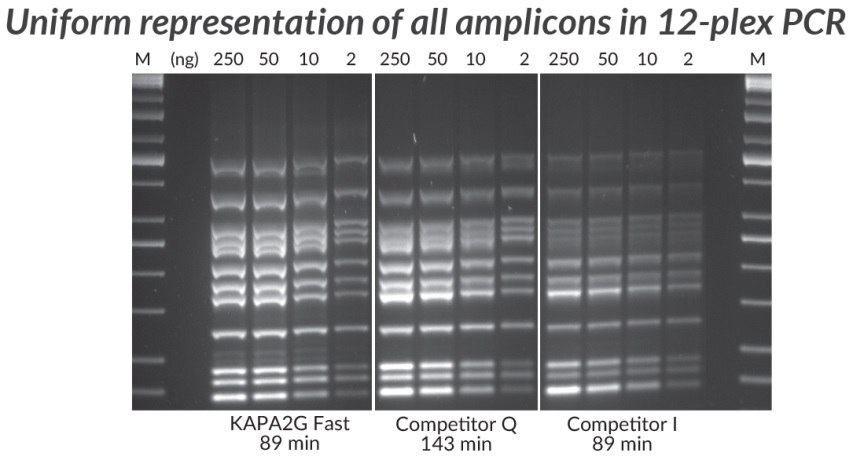
Figure 4.*Multiplex PCR (12-plex) performed with the KAPA2G Fast Multiplex PCR Kit, Competitor Q, and Competitor I. Achieving uniform representation of all amplicons in a complex multiplex assay is a challenge due to amplification bias— a result of differences in amplicon length, secondary structure, and priming efficiency. Reactions (25 μL) contained 1X PCR Master Mix (KAPA and Competitor Q) or 1X PCR Buffer, 3 mM MgCl2, 0.2 mM of each dNTP and 1 U of hot-start Taq DNA Polymerase (home brew multiplex reagents, with Competitor I). Human genomic DNA was used as template (250 – 2 ng per reaction), and primers were supplied at 0.2 μM each. Cycling was performed according to manufacturers’ recommendations (30 cycles).
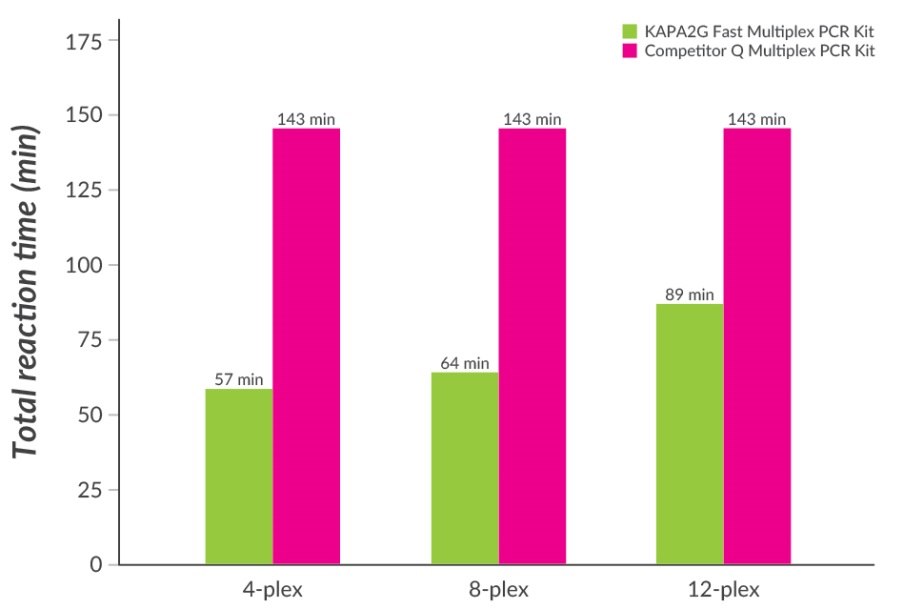
Figure 5.Fast Multiplex PCR with KAPA2G Fast Multiplex PCR Kits. Multiplex PCR with wild-type Taq typically requires long annealing and extension times to allow primer annealing and extension of all primers in the multiplex. Total PCR cycling times required for 4-plex, 8-plex, and 12-plex multiplex PCRs (30 cycles, set up according to the manufacturers’ recommendations) with KAPA2G Fast Multiplex PCR Kits and Competitor Q Multiplex PCR Kit (which contains wildtype Taq DNA polymerase) are shown. Time savings of 40 – 60% are possible with the KAPA2G Fast Multiplex PCR Kit.
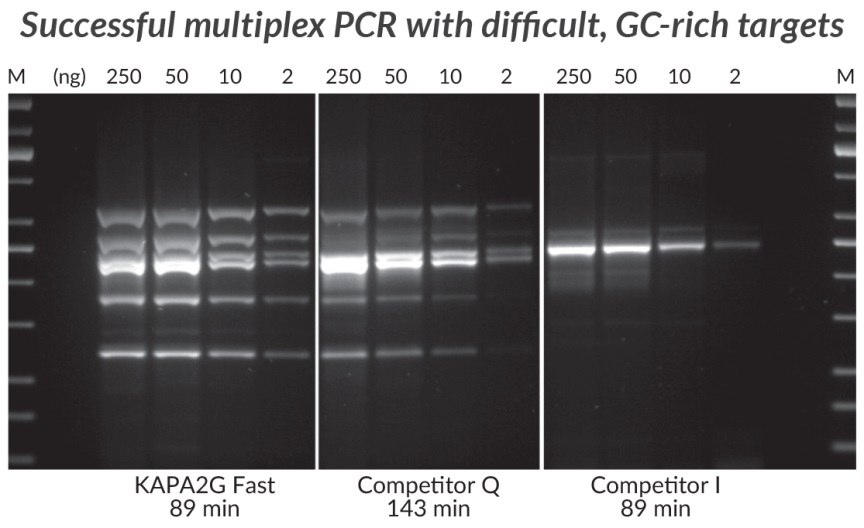
Figure 6.GC-rich Multiplex PCR (6-plex) performed with the KAPA2G Fast Multiplex PCR Kit, Competitor Q and Competitor I. Successful Multiplex PCR with wild-type Taq is limited to easy, simple targets that can be amplified with equal efficiency. The improved processivity of the engineered KAPA2G Fast DNA Polymerase allows uniform multiplex PCR of a broad range of difficult targets. Reactions (25 μL) contained 1X PCR Master Mix (KAPA and Competitor Q) or 1X PCR Buffer, 3 mM MgCl2, 0.2 mM of each dNTP and 1 U of hot start Taq DNA Polymerase (home brew multiplex reagents, with Competitor I). Human genomic DNA was used as template (250 – 2 ng per reaction), and primers were supplied at 0.2 μM each. DMSO (5%) and KAPA Enhancer 1 (1X) was added to all reactions. Cycling was performed according to manufacturers’ recommendations (30 cycles). Amplicons range in size from 241 – 642 bp, and in GC content from 72.7 – 83.8%.
KAPA2G ROBUST DNA POLYMERASE
KAPA2G Robust DNA Polymerase was evolved to solve inconsistent amplification across a broad range of amplicon types (GC- and AT-rich). This product enables higher processivity and specific activity, which translates to robust PCR performance, high sensitivity, and improved tolerance to common inhibitors. This high-performance kit is ideally suited for challenging PCR applications and difficult samples, eliminating the need for optimization using multiple enzymes and protocols.
KAPA2G Robust DNA Polymerase provides:
- Applications requiring higher yield per unit enzyme
- Colony PCR
- Tolerance to inhibitor carryover and crude sample pcr (e.g., FFPE)
- Routine PCR using the HotStart or Readymix formulation
Efficient Amplification of GC- and AT-rich Targets:
- Robust performance across a wide range of GC- and AT-rich templates
- Increased PCR success rates
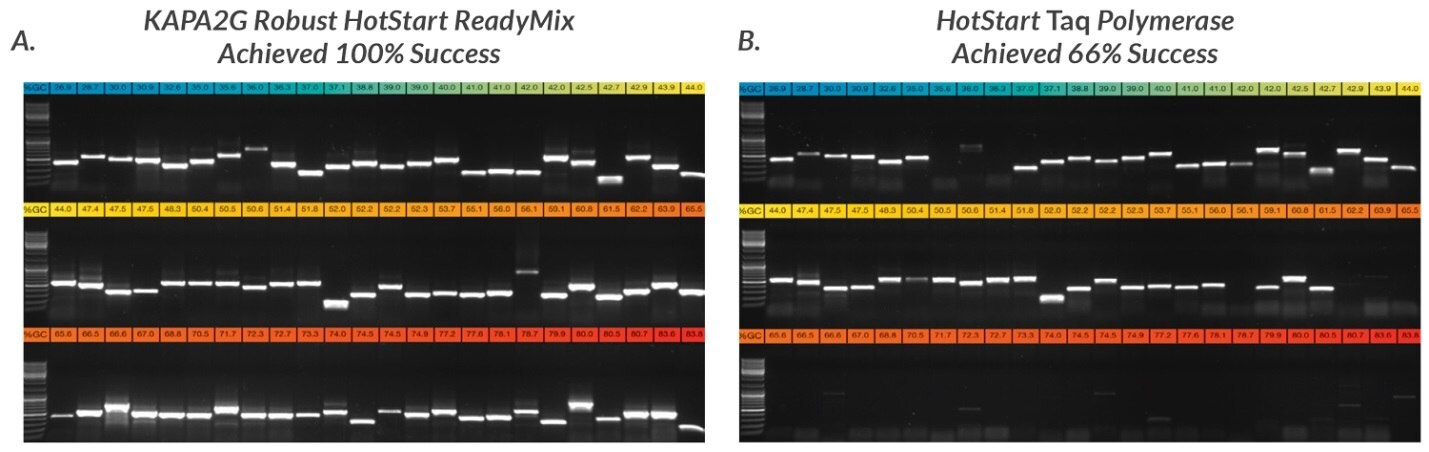
Figure 7.Half of each of the PCR products obtained with 72 of the 96 primer sets used in this study were electrophoresed in a 1% TBE-agarose gel. Amplicons were loaded in order of increasing GC content, with the lowest GC content (27%, blue) at the top left hand side and the highest GC content (84%, red) at the bottom right hand side of each composite gel image. Primers selected for this study had variable primer lengths, sequence composition, theoretical melting temperatures and other design features. Some primers contained 5'-tails for post-PCR sequencing using M13 or other standard sequencing primers. A. KAPA2G Robust HotStart ReadyMix reactions (25 μL) were performed as outlined in the User Guide. B. Wild-type Taq reactions (25 μL, containing 0.5 U Taq per reaction) were performed in Taq reaction buffer (1.5 mM MgCl2 at 1X), using the same final primer and dNTP concentrations as for KAPA2G Robust.
All reactions contained 25 ng human genomic DNA. 5% DMSO was included in all reactions (KAPA2G Robust and Taq) targeting amplicons with a GC content >70%.
Improved Tolerance to Common PCR Inhibitors
- Efficient amplification from crude samples
- Higher yield and sensitivity per unit of enzyme
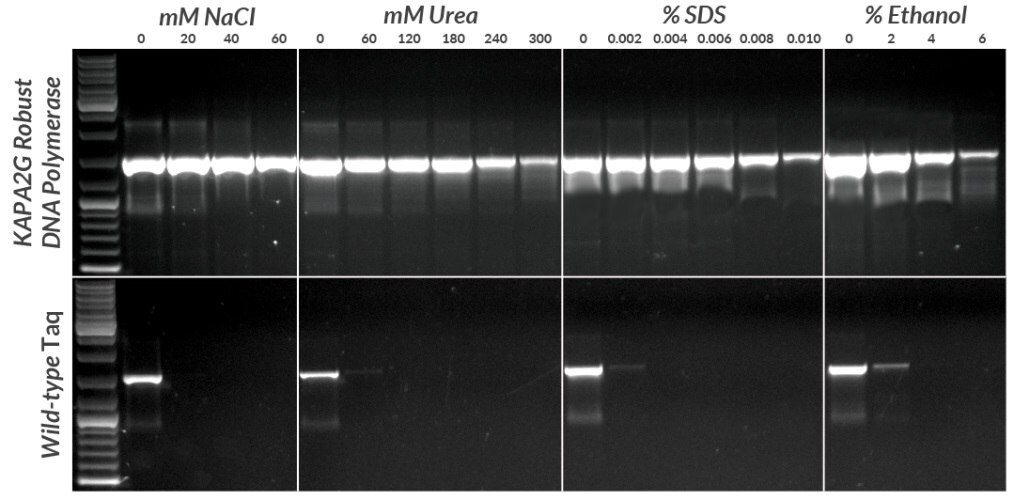
Figure 8.Amplification of a 1.5 kb fragment from 1 pg plasmid DNA in the presence of four common PCR inhibitors using the KAPA2G Robust HotStart PCR Kit and wild-type hot start Taq polymerase. All reactions contained 0.5 units of enzyme per 25 μL reaction. KAPA2G Robust HotStart Buffer B was used throughout, with the addition of KAPAEnhancer 1 for reactions containing SDS. Cycling was performed with an Eppendorf Mastercycler epgradient S, using a standard 3-step cycling profile (35 cycles) with an annealing temperature of 64ºC and 1.5 min extension time per cycle for all enzymes.
Unrivalled Performance in Colony PCR
- Higher yields and improved consistency of PCR direct from E. coli and yeast cells
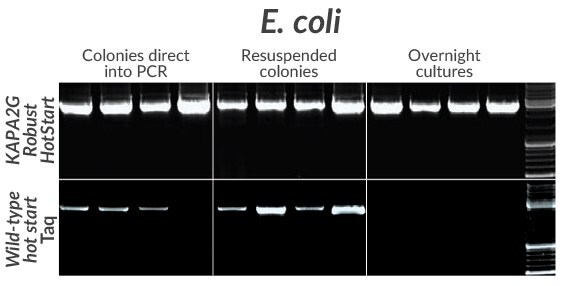
Figure 9.Amplification of a cloned 2.7 kb insert from four commonly used E. coli strains (DH5a, DH10B, JM109, or BL21) using KAPA2G Robust HotStart (top) or wildtype Taq (bottom). Colonies (grown on LB-agar + Amp plates) were either resuspended directly in individual PCR reactions (left) or first resuspended in PCR grade water and then added to PCR reaction mixes (middle). For overnight cultures (prepared in LB + Amp), 1 μ L was added directly to the PCR mix (right).
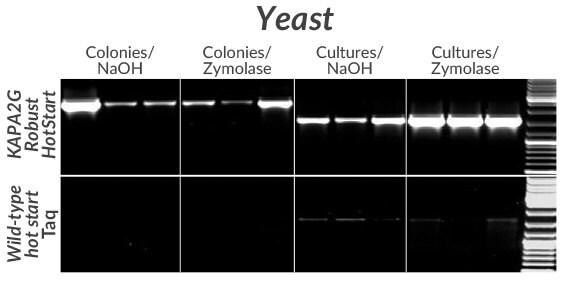
Figure 10.Amplification of a 2.5 kb (left) or 1.6 kb (right) fragment from the GSH1 gene from three commonly used S. cerevisiae strains (BY4742, FY23, and W303) using KAPA2G Robust HotStart (top) or wildtype Taq (bottom). Colonies (from YPD-agar plates) or YPD overnight cultures were first lysed in 50 μ L volumes with NaOH or Zymolase (as indicated).
KAPA LONG RANGE PCR SYSTEM
The KAPA Long Range PCR system is a blend of Taq DNA polymerase and an engineered archaeal (B-family) DNA polymerase possessing proofreading capability. This two-enzyme system is designed specifically to support long-range and/or sensitive PCR, and possesses 3 – 4X higher fidelity than Taq polymerase. In the hotstart formulation, KAPA Long Range is combined with a proprietary antibody that inactivates the enzyme until the first denaturation step, eliminating spurious amplification products and increasing reaction efficiency and sensitivity.
The KAPA Long Range PCR kits are ideally suited for:
- PCR amplification of long targets and/or PCR using low amounts of template DNA
- Standard short- and mid-range PCR amplification
- Production of PCR products to be used for ligation into 3'-T-overhang cloning vectors
Benefits include:
- Amplification of targets with high sensitivity and specificity up to 15 kb
- High yields with low input DNA
- Exhibits high yield, greater sensitivity, and specificity where competitor long-range systems fail*
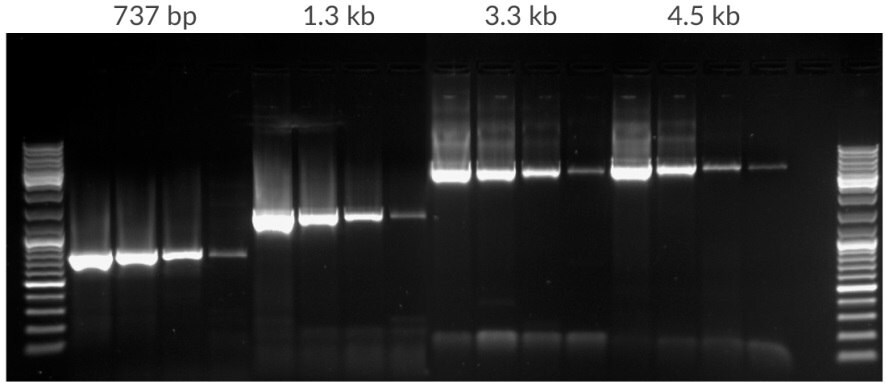
Figure 11.Amplification of template dilution of human genomic DNA starting with 50 ng, 10 ng, 2 ng, 400 pg per 25 μL. Amplicons were 737 bp, 1.3 kb, 3.3 kb, and 4.5 kb in lengths. 35 cycles, 72ºC extension temperature.
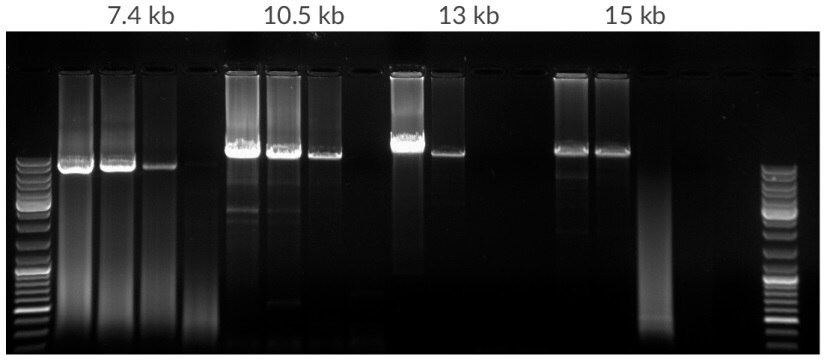
Figure 12.Amplification of template dilution of human genomic DNA starting with 50 ng, 10 ng, 2 ng, 400 pg per 25 μL. Amplicons were 7.5 kb, 10.5 kb, 13 kb, and 15 kb in lengths. 35 cycles, 68ºC extension temperature.
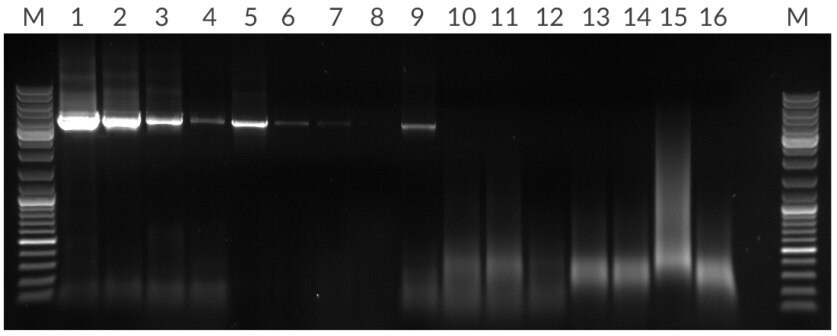
Figure 13.*Amplification of 4.5 kb fragment from 50 ng, 10 ng, 2 ng, and 400 pg of human genomic DNA per 25 μL reaction. Lanes: (M) Marker, (1 – 4) KAPA Long Range HotStart, (5 – 8) Competitor T, (13 – 16) Competitor R.
KAPA MOUSE GENOTYPING
Go from tail to type in 1 hour.
KAPA Mouse Genotyping Kits are designed for the reliable extraction and amplification of DNA fragments from mouse tissue in as little as 1 hour, as compared to ≥1 day with conventional protocols. The kits include KAPA Express Extract, a novel thermostable protease and buffer system that allows for the extraction of PCR-ready DNA from mouse tissue in as little as 15 minutes, and KAPA2G Fast Genotyping Mix with dye, containing a DNA polymerase engineered via directed evolution for high processivity and extreme speed.
KAPA Mouse Genotyping kits provide:
- 15-minute extraction eliminates organic solvents, pH neutralization and centrifugation
- 45-minute amplification with high processivity and extreme speed
- Convenient master mix format with loading dye in both hot start and non-hot start formulations
Outperforms Crude Extraction Methods and Commercial Kits
- Increased turnaround times and specificity
- Decreased cost

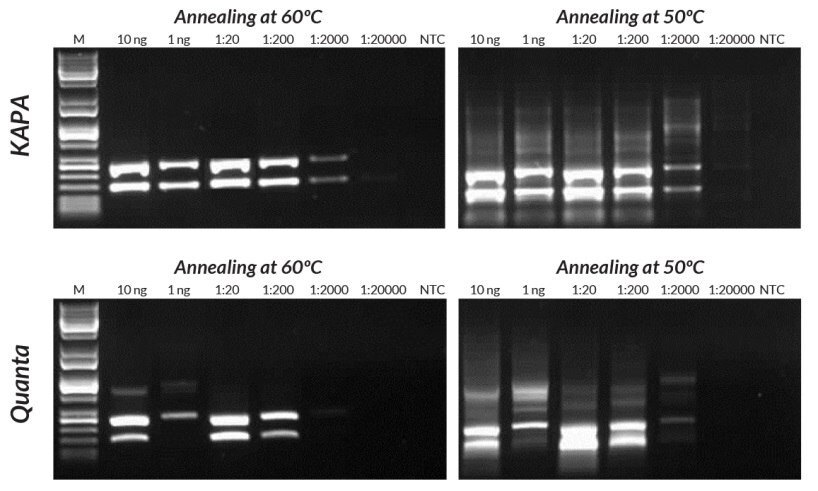
Figures 14 and 15.Compared to the Quanta Mouse Genotyping Kit, KAPA Mouse Genotyping Kits exhibit better sensitivity, yield, and specificity in multiplex PCR. Reactions were performed with annealing at either 50°C or 60°C to demonstrate the effect of using sub-optimal annealing temperatures in genotyping assays.
Increased Throughput, Turnaround Time, and Reliability
- PCR-ready DNA generated in as little as 15 minutes
- Minimal handling, thereby reducing the risk of sample loss or contamination
- Sufficient template for multiple assays; easily scaled to handle samples in a 96-well format
- Fast and reliable amplification of DNA fragments across a wide range of amplicon lengths and GC contents
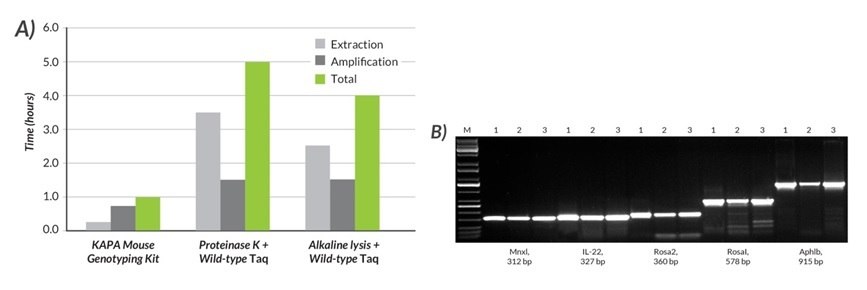
Figures 16 and 17.A) The total time required for DNA extraction and amplification was compared using the KAPA Mouse Genotyping Kit, Proteinase K and wild-type Taq, and alkaline lysis and wild-type Taq. B) Results for five amplicons (312 – 915 bp) generated with the KAPA Mouse Genotyping Kit (1) were compared to those obtained with two commonly used methods (2 and 3). With the KAPA Mouse Genotyping Kit, DNA lysates were prepared from mouse tails with the rapid (15 min), single-tube KAPA Express Extract system. Amplification with the KAPA2G Fast HotStart ReadyMix with dye was completed in 45 min. In contrast, DNA lysates were prepared with a ~3.5-hr Proteinase K protocol (2) or a rapid (~2.5 hr) alkaline lysis method (3). In both cases, amplification was performed with wild-type Taq (1.5 hr cycling protocol). Results obtained with the KAPA Mouse Genotyping Kit were equal or better (more specific) than those obtained with other methods, which (depending on the exact DNA extraction protocol used), may take at least four times as long, or up to 1 day to complete.
KAPA EXPRESS EXTRACT
Rapid and Efficient DNA Extraction
KAPA Express Extract is a novel thermostable protease and buffer system that allows for the extraction of PCR-ready DNA in as little as 15 minutes. DNA extractions are conveniently performed in a single-tube, greatly reducing the risk of sample loss and contamination. The combination of KAPA Express Extract and KAPA2G Robust HotStart ReadyMix provides a high performance solution for rapid DNA extraction and consistent downstream amplification.
KAPA Express Extract provides:
- A rapid extraction protocol with PCR-ready DNA in 15 minutes for consistent downstream amplification of challenging sample types
- Versatile, optimized kit for successful DNA extraction from a variety of samples and tissue types, including buccal swabs, hair follicles, fish tissue, and bird feathers
- Single-tube reaction minimizes risk of contamination
- Efficient extraction from “Guthrie”cards, FTA® Elute cards, and FTA cards

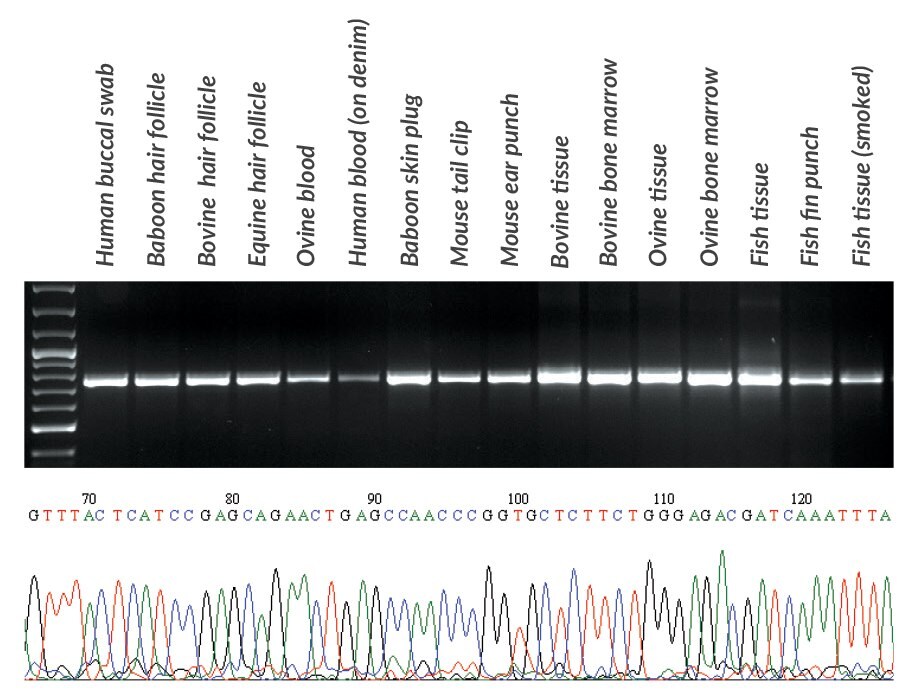
Figures 18 and 19.DNA was extracted with KAPA Express Extract from various samples obtained from mammals and fish. From each extract, 2 μL was used directly (without quantification) in a PCR containing KAPA2G Robust HotStart ReadyMix and primers for the ~650 bp cytochrome c oxidase I gene fragment commonly used in species identification (Ivanova, et al., 2007). PCR products (10 μL) were analyzed in a 1% agarose gel. Sample origin and type is displayed above the gel. Reaction products were used directly in standard Sanger sequencing reactions using out-nested M13 primers (2 μL PCR product per 10 μL sequencing reaction). Sequence data was of a high quality and enabled the identification of each species. A section of the sequence trace from Seriola lalandi (Yellowtail amberjack) tissue is presented in the bottom panel.
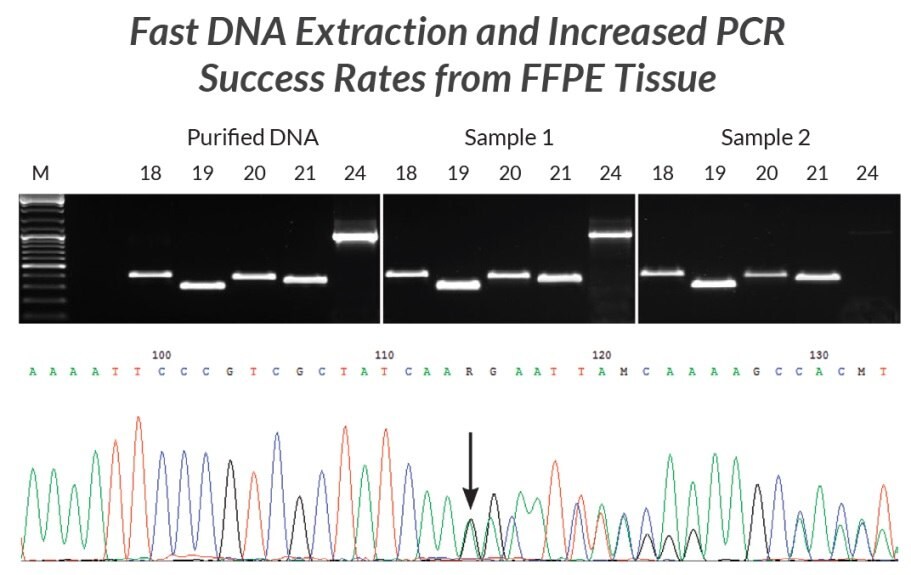
Figure 20.DNA extracts were prepared from two different FFPE samples using KAPA Express Extract. Each extract was used directly (without quantification) in multiple PCRs containing KAPA2G Robust HotStart ReadyMix and primers for five different fragments (293 bp – 1 kb) of the EGFR gene (corresponding to exons 18 – 21 and 24). Results were compared to those obtained using the same reaction and cycling conditions but using 1 ng purified human genomic DNA as template. With the exception of the 1 kb exon 24 fragment from the older sample, yields and reaction efficiencies were comparable between the FFPE DNA extracts and purified genomic DNA. The PCR products generated from sample 1 were diluted 1:10 and used directly in standard Sanger sequencing reactions. Sequence data (bottom panel, Sample 1 exon 19 fragment) was of a high quality. The mixed sequence starting at the position marked with the arrow confirmed the presence of a 15-nt deletion associated with non-small cell carcinoma diagnosed in the patient from whom Sample 1 was collected.
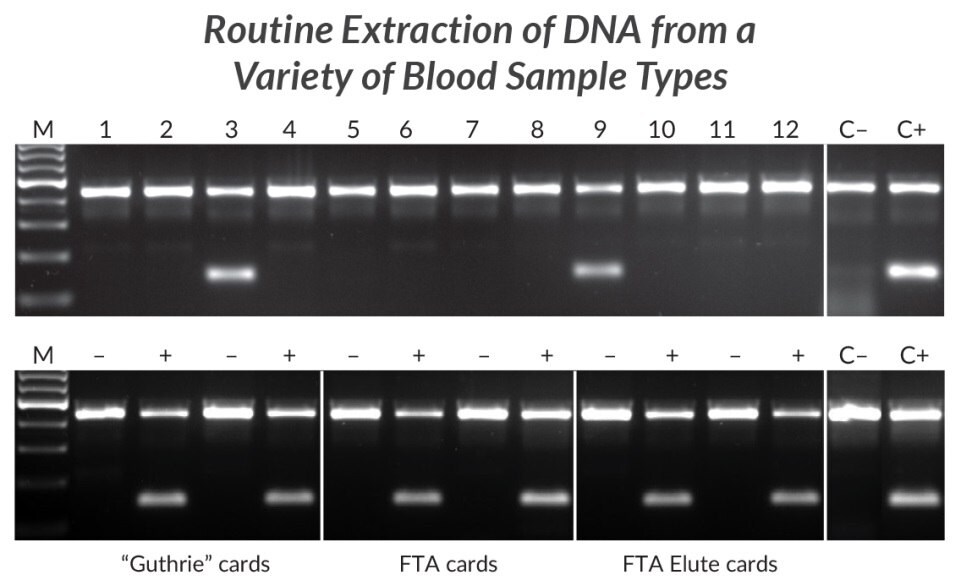
Figure 21.Extraction and amplification of DNA from different blood sample types for detection of the HLA-B*27 allele. DNA was extracted from 12 human EDTA blood samples with KAPA Express Extract (top panel). 2 μL of each extract was added directly to a 25 μL PCR containing KAPA2G Robust HotStart ReadyMix and two primer sets. The internal control primer set targets a 429 bp fragment of the beta globin gene, whereas the second primer set targets a 141 bp fragment of the HLA-B*27 locus in a sequence-specific manner. Two of the 12 individuals tested positive for the HLA-B*27 allele associated with ankylosing spondylitis. Lanes C- and C+ represent HLA-B*27 negative and positive controls respectively (1 ng purified human genomic DNA as template). DNA was extracted from “Guthrie” cards, FTA cards, or FTA Elute cards (bottom panel) spotted with blood of individuals confirmed to be HLA-B*27 positive (+) or negative (-). DNA extraction and amplification conditions and controls (C- and C+) were the same as for the top panel.
KAPA3G PLANT PCR KITS
KAPA3G Plant PCR Kits are designed to utilize either purified DNA or crude extracted DNA for PCR of plant-derived DNA. The kit contains a KAPA3G DNA polymerase, which is engineered for improved tolerance to common plant-derived PCR inhibitors such as polyphenolics and polysaccharides.
KAPA3G Plant PCR Kits provide:
- Fast PCR direct from plant tissues such as leaf discs, seeds and crude plant extracts
- Streamlined workflows for transgenic screening
- Improved PCR success rates and reproducibility
Streamline Workflows and Improve Turnaround Times
- Perform PCR in half the time compared with wild-type enzymes
- Eliminate the need for time-consuming DNA extractions
- Suitable for direct PCR
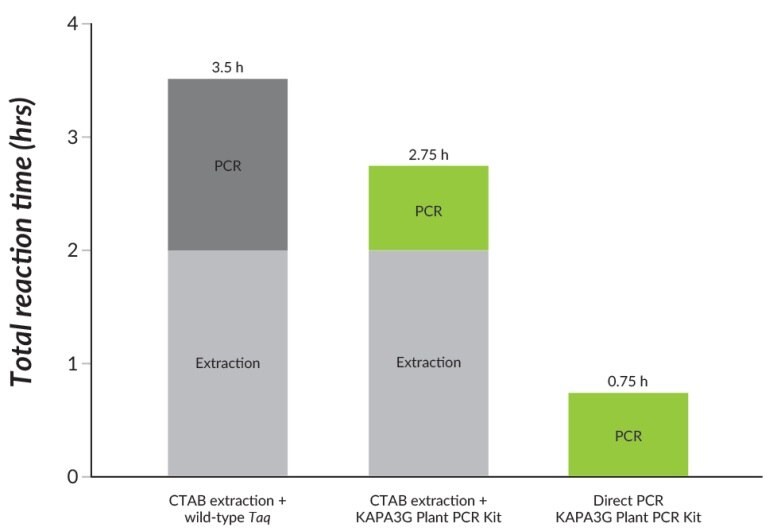
Figure 22.Direct PCR using the KAPA3G Plant PCR Kit outperforms CTAB extraction and standard PCR using wild-type Taq, in significantly shorter turnaround times.
Amplify Long Targets from Crude Samples and Purified DNA
- Amplify fragments up to 5 kb
- High yield and specificity
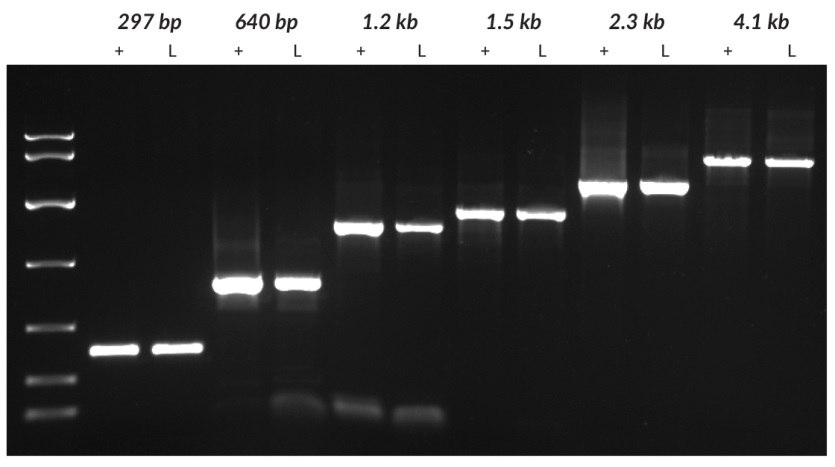
Figure 23.argets of different lengths (297 bp and 4100 bp from tobacco, 640 bp from tomato, 1221 bp from grapevine, and 1448 bp and 2249 bp from potato) were amplified from purified DNA (+) or leaf discs (L) using the KAPA3G Plant PCR Kit. For each species, genomic DNA was purified using a commercial DNA purification kit. Crude material was sampled using a 0.5 mm diameter Harris Uni-Core™ sampling tool. Purified DNA (1 – 10 ng per reaction, depending on species) and crude samples were used as templates in 50 μL PCRs, with 40 cycles of amplification. Reaction products were analyzed on a 1% agarose gel. KAPA Express DNA Ladder (100, 200, 400, 800, 1600, 4000, 8000 bp) was used as a MW marker.
Perform Direct PCR from a Variety of Plant Species and Tissue Types
- Direct PCR with leaf disc or seed as template
- No need for time-consuming DNA extractions
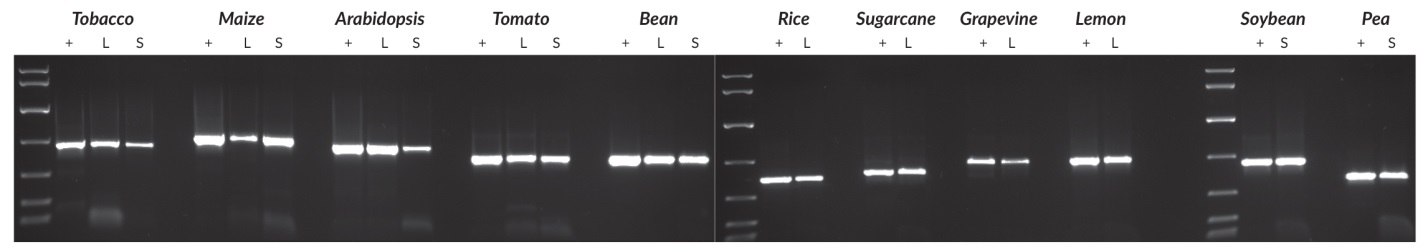
Figure 24.Plant genomic DNA was purified from all species using a commercial DNA purification kit. A Harris Uni-Core™ sampling tool (0.5 mm diameter) was used to sample leaves (all species) or seeds (all species except tobacco and Arabidopsis; for these, one crushed seed was used per reaction). PCRs (50 μL) contained the crude sample or 1 – 10 ng purified DNA (depending on the species), and 40 cycles were performed in all cases. Targets ranged between 500 and 900 bp, and reaction products were analyzed in a 1% agarose gel. KAPA Express DNA Ladder (100, 200, 400, 800, 1600, 4000, 8000 bp) was used as a MW marker.
Improve Success Rates with Novel Crude Sample Plant PCR Workflow
- Use extraction buffer to prepare crude extracts for plant PCR in just 5 minutes
- High success rates with even the most challenging sample types
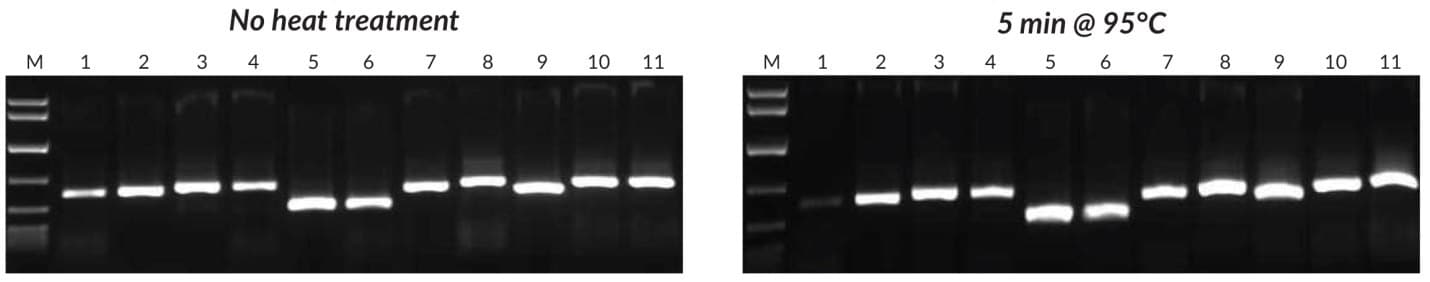
Figure 25.Crude sample PCR with the Tab c/d primer pair was performed on leaves and/or seeds from 8 plants, with 1 μL of crude extract prepared without heat treatment (left) or 1 μL of crude extract prepared with heat treatment (right). Reactions were set up as described in the KAPA3G Plant PCR Kit TDS, and 45 cycles of PCR performed with annealing at 55°C, and 20 sec extension at 72°C per cycle. KAPA Express DNA Ladder (100, 200, 400, 800, 1600, 4000, 8000 bp) was used as a MW marker. 1: Eucalyptus; 2: Grapevine; 3: Wheat leaf; 4: Wheat seed; 5: Canola leaf; 6: Canola seed; 7: Rice seed; 8: Barley seed; 9: Corn kernel; 10: Cotton seed; 11: Cotton leaf.
如要继续阅读,请登录或创建帐户。
暂无帐户?